- Affiliate Program

- UNITED STATES
- 台灣 (TAIWAN)
- TÜRKIYE (TURKEY)
- Academic Editing Services
- - Research Paper
- - Journal Manuscript
- - Dissertation
- - College & University Assignments
- Admissions Editing Services
- - Application Essay
- - Personal Statement
- - Recommendation Letter
- - Cover Letter
- - CV/Resume
- Business Editing Services
- - Business Documents
- - Report & Brochure
- - Website & Blog
- Writer Editing Services
- - Script & Screenplay
- Our Editors
- Client Reviews
- Editing & Proofreading Prices
- Wordvice Points
- Partner Discount
- Plagiarism Checker
- APA Citation Generator
- MLA Citation Generator
- Chicago Citation Generator
- Vancouver Citation Generator
- - APA Style
- - MLA Style
- - Chicago Style
- - Vancouver Style
- Writing & Editing Guide
- Academic Resources
- Admissions Resources

How to Write the Results/Findings Section in Research
What is the research paper Results section and what does it do?
The Results section of a scientific research paper represents the core findings of a study derived from the methods applied to gather and analyze information. It presents these findings in a logical sequence without bias or interpretation from the author, setting up the reader for later interpretation and evaluation in the Discussion section. A major purpose of the Results section is to break down the data into sentences that show its significance to the research question(s).
The Results section appears third in the section sequence in most scientific papers. It follows the presentation of the Methods and Materials and is presented before the Discussion section —although the Results and Discussion are presented together in many journals. This section answers the basic question “What did you find in your research?”
What is included in the Results section?
The Results section should include the findings of your study and ONLY the findings of your study. The findings include:
- Data presented in tables, charts, graphs, and other figures (may be placed into the text or on separate pages at the end of the manuscript)
- A contextual analysis of this data explaining its meaning in sentence form
- All data that corresponds to the central research question(s)
- All secondary findings (secondary outcomes, subgroup analyses, etc.)
If the scope of the study is broad, or if you studied a variety of variables, or if the methodology used yields a wide range of different results, the author should present only those results that are most relevant to the research question stated in the Introduction section .
As a general rule, any information that does not present the direct findings or outcome of the study should be left out of this section. Unless the journal requests that authors combine the Results and Discussion sections, explanations and interpretations should be omitted from the Results.
How are the results organized?
The best way to organize your Results section is “logically.” One logical and clear method of organizing research results is to provide them alongside the research questions—within each research question, present the type of data that addresses that research question.
Let’s look at an example. Your research question is based on a survey among patients who were treated at a hospital and received postoperative care. Let’s say your first research question is:
“What do hospital patients over age 55 think about postoperative care?”
This can actually be represented as a heading within your Results section, though it might be presented as a statement rather than a question:
Attitudes towards postoperative care in patients over the age of 55
Now present the results that address this specific research question first. In this case, perhaps a table illustrating data from a survey. Likert items can be included in this example. Tables can also present standard deviations, probabilities, correlation matrices, etc.
Following this, present a content analysis, in words, of one end of the spectrum of the survey or data table. In our example case, start with the POSITIVE survey responses regarding postoperative care, using descriptive phrases. For example:
“Sixty-five percent of patients over 55 responded positively to the question “ Are you satisfied with your hospital’s postoperative care ?” (Fig. 2)
Include other results such as subcategory analyses. The amount of textual description used will depend on how much interpretation of tables and figures is necessary and how many examples the reader needs in order to understand the significance of your research findings.
Next, present a content analysis of another part of the spectrum of the same research question, perhaps the NEGATIVE or NEUTRAL responses to the survey. For instance:
“As Figure 1 shows, 15 out of 60 patients in Group A responded negatively to Question 2.”
After you have assessed the data in one figure and explained it sufficiently, move on to your next research question. For example:
“How does patient satisfaction correspond to in-hospital improvements made to postoperative care?”
This kind of data may be presented through a figure or set of figures (for instance, a paired T-test table).
Explain the data you present, here in a table, with a concise content analysis:
“The p-value for the comparison between the before and after groups of patients was .03% (Fig. 2), indicating that the greater the dissatisfaction among patients, the more frequent the improvements that were made to postoperative care.”
Let’s examine another example of a Results section from a study on plant tolerance to heavy metal stress . In the Introduction section, the aims of the study are presented as “determining the physiological and morphological responses of Allium cepa L. towards increased cadmium toxicity” and “evaluating its potential to accumulate the metal and its associated environmental consequences.” The Results section presents data showing how these aims are achieved in tables alongside a content analysis, beginning with an overview of the findings:
“Cadmium caused inhibition of root and leave elongation, with increasing effects at higher exposure doses (Fig. 1a-c).”
The figure containing this data is cited in parentheses. Note that this author has combined three graphs into one single figure. Separating the data into separate graphs focusing on specific aspects makes it easier for the reader to assess the findings, and consolidating this information into one figure saves space and makes it easy to locate the most relevant results.
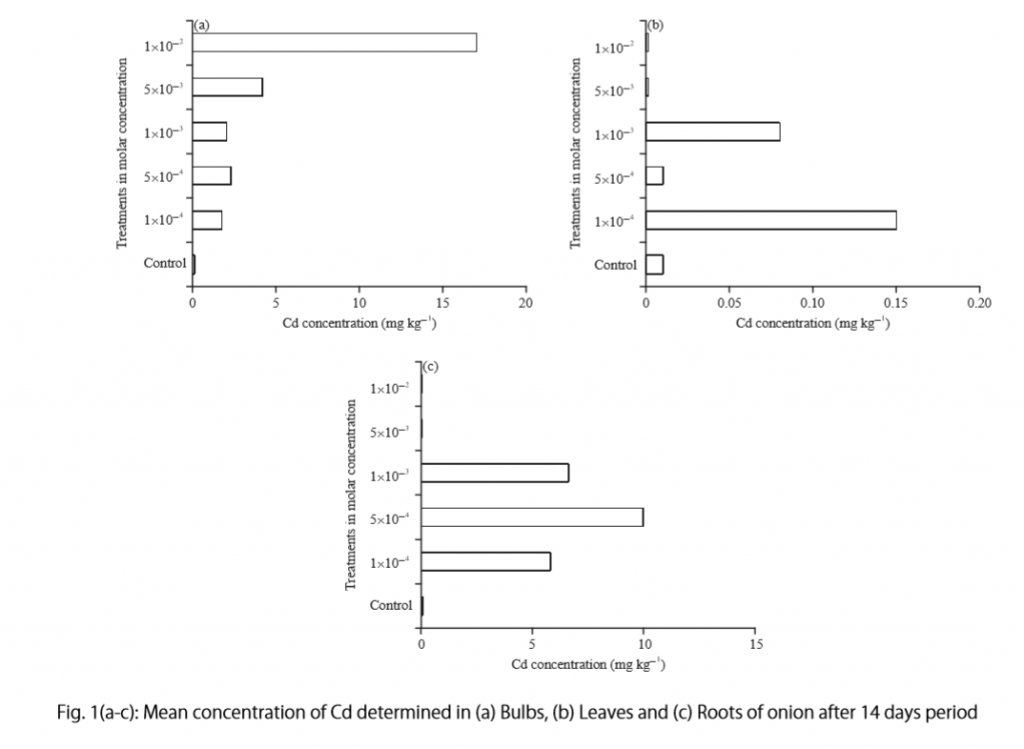
Following this overall summary, the relevant data in the tables is broken down into greater detail in text form in the Results section.
- “Results on the bio-accumulation of cadmium were found to be the highest (17.5 mg kgG1) in the bulb, when the concentration of cadmium in the solution was 1×10G2 M and lowest (0.11 mg kgG1) in the leaves when the concentration was 1×10G3 M.”
Captioning and Referencing Tables and Figures
Tables and figures are central components of your Results section and you need to carefully think about the most effective way to use graphs and tables to present your findings . Therefore, it is crucial to know how to write strong figure captions and to refer to them within the text of the Results section.
The most important advice one can give here as well as throughout the paper is to check the requirements and standards of the journal to which you are submitting your work. Every journal has its own design and layout standards, which you can find in the author instructions on the target journal’s website. Perusing a journal’s published articles will also give you an idea of the proper number, size, and complexity of your figures.
Regardless of which format you use, the figures should be placed in the order they are referenced in the Results section and be as clear and easy to understand as possible. If there are multiple variables being considered (within one or more research questions), it can be a good idea to split these up into separate figures. Subsequently, these can be referenced and analyzed under separate headings and paragraphs in the text.
To create a caption, consider the research question being asked and change it into a phrase. For instance, if one question is “Which color did participants choose?”, the caption might be “Color choice by participant group.” Or in our last research paper example, where the question was “What is the concentration of cadmium in different parts of the onion after 14 days?” the caption reads:
“Fig. 1(a-c): Mean concentration of Cd determined in (a) bulbs, (b) leaves, and (c) roots of onions after a 14-day period.”
Steps for Composing the Results Section
Because each study is unique, there is no one-size-fits-all approach when it comes to designing a strategy for structuring and writing the section of a research paper where findings are presented. The content and layout of this section will be determined by the specific area of research, the design of the study and its particular methodologies, and the guidelines of the target journal and its editors. However, the following steps can be used to compose the results of most scientific research studies and are essential for researchers who are new to preparing a manuscript for publication or who need a reminder of how to construct the Results section.
Step 1 : Consult the guidelines or instructions that the target journal or publisher provides authors and read research papers it has published, especially those with similar topics, methods, or results to your study.
- The guidelines will generally outline specific requirements for the results or findings section, and the published articles will provide sound examples of successful approaches.
- Note length limitations on restrictions on content. For instance, while many journals require the Results and Discussion sections to be separate, others do not—qualitative research papers often include results and interpretations in the same section (“Results and Discussion”).
- Reading the aims and scope in the journal’s “ guide for authors ” section and understanding the interests of its readers will be invaluable in preparing to write the Results section.
Step 2 : Consider your research results in relation to the journal’s requirements and catalogue your results.
- Focus on experimental results and other findings that are especially relevant to your research questions and objectives and include them even if they are unexpected or do not support your ideas and hypotheses.
- Catalogue your findings—use subheadings to streamline and clarify your report. This will help you avoid excessive and peripheral details as you write and also help your reader understand and remember your findings. Create appendices that might interest specialists but prove too long or distracting for other readers.
- Decide how you will structure of your results. You might match the order of the research questions and hypotheses to your results, or you could arrange them according to the order presented in the Methods section. A chronological order or even a hierarchy of importance or meaningful grouping of main themes or categories might prove effective. Consider your audience, evidence, and most importantly, the objectives of your research when choosing a structure for presenting your findings.
Step 3 : Design figures and tables to present and illustrate your data.
- Tables and figures should be numbered according to the order in which they are mentioned in the main text of the paper.
- Information in figures should be relatively self-explanatory (with the aid of captions), and their design should include all definitions and other information necessary for readers to understand the findings without reading all of the text.
- Use tables and figures as a focal point to tell a clear and informative story about your research and avoid repeating information. But remember that while figures clarify and enhance the text, they cannot replace it.
Step 4 : Draft your Results section using the findings and figures you have organized.
- The goal is to communicate this complex information as clearly and precisely as possible; precise and compact phrases and sentences are most effective.
- In the opening paragraph of this section, restate your research questions or aims to focus the reader’s attention to what the results are trying to show. It is also a good idea to summarize key findings at the end of this section to create a logical transition to the interpretation and discussion that follows.
- Try to write in the past tense and the active voice to relay the findings since the research has already been done and the agent is usually clear. This will ensure that your explanations are also clear and logical.
- Make sure that any specialized terminology or abbreviation you have used here has been defined and clarified in the Introduction section .
Step 5 : Review your draft; edit and revise until it reports results exactly as you would like to have them reported to your readers.
- Double-check the accuracy and consistency of all the data, as well as all of the visual elements included.
- Read your draft aloud to catch language errors (grammar, spelling, and mechanics), awkward phrases, and missing transitions.
- Ensure that your results are presented in the best order to focus on objectives and prepare readers for interpretations, valuations, and recommendations in the Discussion section . Look back over the paper’s Introduction and background while anticipating the Discussion and Conclusion sections to ensure that the presentation of your results is consistent and effective.
- Consider seeking additional guidance on your paper. Find additional readers to look over your Results section and see if it can be improved in any way. Peers, professors, or qualified experts can provide valuable insights.
One excellent option is to use a professional English proofreading and editing service such as Wordvice, including our paper editing service . With hundreds of qualified editors from dozens of scientific fields, Wordvice has helped thousands of authors revise their manuscripts and get accepted into their target journals. Read more about the proofreading and editing process before proceeding with getting academic editing services and manuscript editing services for your manuscript.
As the representation of your study’s data output, the Results section presents the core information in your research paper. By writing with clarity and conciseness and by highlighting and explaining the crucial findings of their study, authors increase the impact and effectiveness of their research manuscripts.
For more articles and videos on writing your research manuscript, visit Wordvice’s Resources page.
Wordvice Resources
- How to Write a Research Paper Introduction
- Which Verb Tenses to Use in a Research Paper
- How to Write an Abstract for a Research Paper
- How to Write a Research Paper Title
- Useful Phrases for Academic Writing
- Common Transition Terms in Academic Papers
- Active and Passive Voice in Research Papers
- 100+ Verbs That Will Make Your Research Writing Amazing
- Tips for Paraphrasing in Research Papers
- Privacy Policy

Home » Research Findings – Types Examples and Writing Guide
Research Findings – Types Examples and Writing Guide
Table of Contents

Research Findings
Definition:
Research findings refer to the results obtained from a study or investigation conducted through a systematic and scientific approach. These findings are the outcomes of the data analysis, interpretation, and evaluation carried out during the research process.
Types of Research Findings
There are two main types of research findings:
Qualitative Findings
Qualitative research is an exploratory research method used to understand the complexities of human behavior and experiences. Qualitative findings are non-numerical and descriptive data that describe the meaning and interpretation of the data collected. Examples of qualitative findings include quotes from participants, themes that emerge from the data, and descriptions of experiences and phenomena.
Quantitative Findings
Quantitative research is a research method that uses numerical data and statistical analysis to measure and quantify a phenomenon or behavior. Quantitative findings include numerical data such as mean, median, and mode, as well as statistical analyses such as t-tests, ANOVA, and regression analysis. These findings are often presented in tables, graphs, or charts.
Both qualitative and quantitative findings are important in research and can provide different insights into a research question or problem. Combining both types of findings can provide a more comprehensive understanding of a phenomenon and improve the validity and reliability of research results.
Parts of Research Findings
Research findings typically consist of several parts, including:
- Introduction: This section provides an overview of the research topic and the purpose of the study.
- Literature Review: This section summarizes previous research studies and findings that are relevant to the current study.
- Methodology : This section describes the research design, methods, and procedures used in the study, including details on the sample, data collection, and data analysis.
- Results : This section presents the findings of the study, including statistical analyses and data visualizations.
- Discussion : This section interprets the results and explains what they mean in relation to the research question(s) and hypotheses. It may also compare and contrast the current findings with previous research studies and explore any implications or limitations of the study.
- Conclusion : This section provides a summary of the key findings and the main conclusions of the study.
- Recommendations: This section suggests areas for further research and potential applications or implications of the study’s findings.
How to Write Research Findings
Writing research findings requires careful planning and attention to detail. Here are some general steps to follow when writing research findings:
- Organize your findings: Before you begin writing, it’s essential to organize your findings logically. Consider creating an outline or a flowchart that outlines the main points you want to make and how they relate to one another.
- Use clear and concise language : When presenting your findings, be sure to use clear and concise language that is easy to understand. Avoid using jargon or technical terms unless they are necessary to convey your meaning.
- Use visual aids : Visual aids such as tables, charts, and graphs can be helpful in presenting your findings. Be sure to label and title your visual aids clearly, and make sure they are easy to read.
- Use headings and subheadings: Using headings and subheadings can help organize your findings and make them easier to read. Make sure your headings and subheadings are clear and descriptive.
- Interpret your findings : When presenting your findings, it’s important to provide some interpretation of what the results mean. This can include discussing how your findings relate to the existing literature, identifying any limitations of your study, and suggesting areas for future research.
- Be precise and accurate : When presenting your findings, be sure to use precise and accurate language. Avoid making generalizations or overstatements and be careful not to misrepresent your data.
- Edit and revise: Once you have written your research findings, be sure to edit and revise them carefully. Check for grammar and spelling errors, make sure your formatting is consistent, and ensure that your writing is clear and concise.
Research Findings Example
Following is a Research Findings Example sample for students:
Title: The Effects of Exercise on Mental Health
Sample : 500 participants, both men and women, between the ages of 18-45.
Methodology : Participants were divided into two groups. The first group engaged in 30 minutes of moderate intensity exercise five times a week for eight weeks. The second group did not exercise during the study period. Participants in both groups completed a questionnaire that assessed their mental health before and after the study period.
Findings : The group that engaged in regular exercise reported a significant improvement in mental health compared to the control group. Specifically, they reported lower levels of anxiety and depression, improved mood, and increased self-esteem.
Conclusion : Regular exercise can have a positive impact on mental health and may be an effective intervention for individuals experiencing symptoms of anxiety or depression.
Applications of Research Findings
Research findings can be applied in various fields to improve processes, products, services, and outcomes. Here are some examples:
- Healthcare : Research findings in medicine and healthcare can be applied to improve patient outcomes, reduce morbidity and mortality rates, and develop new treatments for various diseases.
- Education : Research findings in education can be used to develop effective teaching methods, improve learning outcomes, and design new educational programs.
- Technology : Research findings in technology can be applied to develop new products, improve existing products, and enhance user experiences.
- Business : Research findings in business can be applied to develop new strategies, improve operations, and increase profitability.
- Public Policy: Research findings can be used to inform public policy decisions on issues such as environmental protection, social welfare, and economic development.
- Social Sciences: Research findings in social sciences can be used to improve understanding of human behavior and social phenomena, inform public policy decisions, and develop interventions to address social issues.
- Agriculture: Research findings in agriculture can be applied to improve crop yields, develop new farming techniques, and enhance food security.
- Sports : Research findings in sports can be applied to improve athlete performance, reduce injuries, and develop new training programs.
When to use Research Findings
Research findings can be used in a variety of situations, depending on the context and the purpose. Here are some examples of when research findings may be useful:
- Decision-making : Research findings can be used to inform decisions in various fields, such as business, education, healthcare, and public policy. For example, a business may use market research findings to make decisions about new product development or marketing strategies.
- Problem-solving : Research findings can be used to solve problems or challenges in various fields, such as healthcare, engineering, and social sciences. For example, medical researchers may use findings from clinical trials to develop new treatments for diseases.
- Policy development : Research findings can be used to inform the development of policies in various fields, such as environmental protection, social welfare, and economic development. For example, policymakers may use research findings to develop policies aimed at reducing greenhouse gas emissions.
- Program evaluation: Research findings can be used to evaluate the effectiveness of programs or interventions in various fields, such as education, healthcare, and social services. For example, educational researchers may use findings from evaluations of educational programs to improve teaching and learning outcomes.
- Innovation: Research findings can be used to inspire or guide innovation in various fields, such as technology and engineering. For example, engineers may use research findings on materials science to develop new and innovative products.
Purpose of Research Findings
The purpose of research findings is to contribute to the knowledge and understanding of a particular topic or issue. Research findings are the result of a systematic and rigorous investigation of a research question or hypothesis, using appropriate research methods and techniques.
The main purposes of research findings are:
- To generate new knowledge : Research findings contribute to the body of knowledge on a particular topic, by adding new information, insights, and understanding to the existing knowledge base.
- To test hypotheses or theories : Research findings can be used to test hypotheses or theories that have been proposed in a particular field or discipline. This helps to determine the validity and reliability of the hypotheses or theories, and to refine or develop new ones.
- To inform practice: Research findings can be used to inform practice in various fields, such as healthcare, education, and business. By identifying best practices and evidence-based interventions, research findings can help practitioners to make informed decisions and improve outcomes.
- To identify gaps in knowledge: Research findings can help to identify gaps in knowledge and understanding of a particular topic, which can then be addressed by further research.
- To contribute to policy development: Research findings can be used to inform policy development in various fields, such as environmental protection, social welfare, and economic development. By providing evidence-based recommendations, research findings can help policymakers to develop effective policies that address societal challenges.
Characteristics of Research Findings
Research findings have several key characteristics that distinguish them from other types of information or knowledge. Here are some of the main characteristics of research findings:
- Objective : Research findings are based on a systematic and rigorous investigation of a research question or hypothesis, using appropriate research methods and techniques. As such, they are generally considered to be more objective and reliable than other types of information.
- Empirical : Research findings are based on empirical evidence, which means that they are derived from observations or measurements of the real world. This gives them a high degree of credibility and validity.
- Generalizable : Research findings are often intended to be generalizable to a larger population or context beyond the specific study. This means that the findings can be applied to other situations or populations with similar characteristics.
- Transparent : Research findings are typically reported in a transparent manner, with a clear description of the research methods and data analysis techniques used. This allows others to assess the credibility and reliability of the findings.
- Peer-reviewed: Research findings are often subject to a rigorous peer-review process, in which experts in the field review the research methods, data analysis, and conclusions of the study. This helps to ensure the validity and reliability of the findings.
- Reproducible : Research findings are often designed to be reproducible, meaning that other researchers can replicate the study using the same methods and obtain similar results. This helps to ensure the validity and reliability of the findings.
Advantages of Research Findings
Research findings have many advantages, which make them valuable sources of knowledge and information. Here are some of the main advantages of research findings:
- Evidence-based: Research findings are based on empirical evidence, which means that they are grounded in data and observations from the real world. This makes them a reliable and credible source of information.
- Inform decision-making: Research findings can be used to inform decision-making in various fields, such as healthcare, education, and business. By identifying best practices and evidence-based interventions, research findings can help practitioners and policymakers to make informed decisions and improve outcomes.
- Identify gaps in knowledge: Research findings can help to identify gaps in knowledge and understanding of a particular topic, which can then be addressed by further research. This contributes to the ongoing development of knowledge in various fields.
- Improve outcomes : Research findings can be used to develop and implement evidence-based practices and interventions, which have been shown to improve outcomes in various fields, such as healthcare, education, and social services.
- Foster innovation: Research findings can inspire or guide innovation in various fields, such as technology and engineering. By providing new information and understanding of a particular topic, research findings can stimulate new ideas and approaches to problem-solving.
- Enhance credibility: Research findings are generally considered to be more credible and reliable than other types of information, as they are based on rigorous research methods and are subject to peer-review processes.
Limitations of Research Findings
While research findings have many advantages, they also have some limitations. Here are some of the main limitations of research findings:
- Limited scope: Research findings are typically based on a particular study or set of studies, which may have a limited scope or focus. This means that they may not be applicable to other contexts or populations.
- Potential for bias : Research findings can be influenced by various sources of bias, such as researcher bias, selection bias, or measurement bias. This can affect the validity and reliability of the findings.
- Ethical considerations: Research findings can raise ethical considerations, particularly in studies involving human subjects. Researchers must ensure that their studies are conducted in an ethical and responsible manner, with appropriate measures to protect the welfare and privacy of participants.
- Time and resource constraints : Research studies can be time-consuming and require significant resources, which can limit the number and scope of studies that are conducted. This can lead to gaps in knowledge or a lack of research on certain topics.
- Complexity: Some research findings can be complex and difficult to interpret, particularly in fields such as science or medicine. This can make it challenging for practitioners and policymakers to apply the findings to their work.
- Lack of generalizability : While research findings are intended to be generalizable to larger populations or contexts, there may be factors that limit their generalizability. For example, cultural or environmental factors may influence how a particular intervention or treatment works in different populations or contexts.
About the author
Muhammad Hassan
Researcher, Academic Writer, Web developer
You may also like

Significance of the Study – Examples and Writing...

Research Design – Types, Methods and Examples

Research Approach – Types Methods and Examples

Dissertation – Format, Example and Template

Research Gap – Types, Examples and How to...

Research Methods – Types, Examples and Guide
- USC Libraries
- Research Guides
Organizing Your Social Sciences Research Paper
- 7. The Results
- Purpose of Guide
- Design Flaws to Avoid
- Independent and Dependent Variables
- Glossary of Research Terms
- Reading Research Effectively
- Narrowing a Topic Idea
- Broadening a Topic Idea
- Extending the Timeliness of a Topic Idea
- Academic Writing Style
- Applying Critical Thinking
- Choosing a Title
- Making an Outline
- Paragraph Development
- Research Process Video Series
- Executive Summary
- The C.A.R.S. Model
- Background Information
- The Research Problem/Question
- Theoretical Framework
- Citation Tracking
- Content Alert Services
- Evaluating Sources
- Primary Sources
- Secondary Sources
- Tiertiary Sources
- Scholarly vs. Popular Publications
- Qualitative Methods
- Quantitative Methods
- Insiderness
- Using Non-Textual Elements
- Limitations of the Study
- Common Grammar Mistakes
- Writing Concisely
- Avoiding Plagiarism
- Footnotes or Endnotes?
- Further Readings
- Generative AI and Writing
- USC Libraries Tutorials and Other Guides
- Bibliography
The results section is where you report the findings of your study based upon the methodology [or methodologies] you applied to gather information. The results section should state the findings of the research arranged in a logical sequence without bias or interpretation. A section describing results should be particularly detailed if your paper includes data generated from your own research.
Annesley, Thomas M. "Show Your Cards: The Results Section and the Poker Game." Clinical Chemistry 56 (July 2010): 1066-1070.
Importance of a Good Results Section
When formulating the results section, it's important to remember that the results of a study do not prove anything . Findings can only confirm or reject the hypothesis underpinning your study. However, the act of articulating the results helps you to understand the problem from within, to break it into pieces, and to view the research problem from various perspectives.
The page length of this section is set by the amount and types of data to be reported . Be concise. Use non-textual elements appropriately, such as figures and tables, to present findings more effectively. In deciding what data to describe in your results section, you must clearly distinguish information that would normally be included in a research paper from any raw data or other content that could be included as an appendix. In general, raw data that has not been summarized should not be included in the main text of your paper unless requested to do so by your professor.
Avoid providing data that is not critical to answering the research question . The background information you described in the introduction section should provide the reader with any additional context or explanation needed to understand the results. A good strategy is to always re-read the background section of your paper after you have written up your results to ensure that the reader has enough context to understand the results [and, later, how you interpreted the results in the discussion section of your paper that follows].
Bavdekar, Sandeep B. and Sneha Chandak. "Results: Unraveling the Findings." Journal of the Association of Physicians of India 63 (September 2015): 44-46; Brett, Paul. "A Genre Analysis of the Results Section of Sociology Articles." English for Specific Speakers 13 (1994): 47-59; Go to English for Specific Purposes on ScienceDirect;Burton, Neil et al. Doing Your Education Research Project . Los Angeles, CA: SAGE, 2008; Results. The Structure, Format, Content, and Style of a Journal-Style Scientific Paper. Department of Biology. Bates College; Kretchmer, Paul. Twelve Steps to Writing an Effective Results Section. San Francisco Edit; "Reporting Findings." In Making Sense of Social Research Malcolm Williams, editor. (London;: SAGE Publications, 2003) pp. 188-207.
Structure and Writing Style
I. Organization and Approach
For most research papers in the social and behavioral sciences, there are two possible ways of organizing the results . Both approaches are appropriate in how you report your findings, but use only one approach.
- Present a synopsis of the results followed by an explanation of key findings . This approach can be used to highlight important findings. For example, you may have noticed an unusual correlation between two variables during the analysis of your findings. It is appropriate to highlight this finding in the results section. However, speculating as to why this correlation exists and offering a hypothesis about what may be happening belongs in the discussion section of your paper.
- Present a result and then explain it, before presenting the next result then explaining it, and so on, then end with an overall synopsis . This is the preferred approach if you have multiple results of equal significance. It is more common in longer papers because it helps the reader to better understand each finding. In this model, it is helpful to provide a brief conclusion that ties each of the findings together and provides a narrative bridge to the discussion section of the your paper.
NOTE: Just as the literature review should be arranged under conceptual categories rather than systematically describing each source, you should also organize your findings under key themes related to addressing the research problem. This can be done under either format noted above [i.e., a thorough explanation of the key results or a sequential, thematic description and explanation of each finding].
II. Content
In general, the content of your results section should include the following:
- Introductory context for understanding the results by restating the research problem underpinning your study . This is useful in re-orientating the reader's focus back to the research problem after having read a review of the literature and your explanation of the methods used for gathering and analyzing information.
- Inclusion of non-textual elements, such as, figures, charts, photos, maps, tables, etc. to further illustrate key findings, if appropriate . Rather than relying entirely on descriptive text, consider how your findings can be presented visually. This is a helpful way of condensing a lot of data into one place that can then be referred to in the text. Consider referring to appendices if there is a lot of non-textual elements.
- A systematic description of your results, highlighting for the reader observations that are most relevant to the topic under investigation . Not all results that emerge from the methodology used to gather information may be related to answering the " So What? " question. Do not confuse observations with interpretations; observations in this context refers to highlighting important findings you discovered through a process of reviewing prior literature and gathering data.
- The page length of your results section is guided by the amount and types of data to be reported . However, focus on findings that are important and related to addressing the research problem. It is not uncommon to have unanticipated results that are not relevant to answering the research question. This is not to say that you don't acknowledge tangential findings and, in fact, can be referred to as areas for further research in the conclusion of your paper. However, spending time in the results section describing tangential findings clutters your overall results section and distracts the reader.
- A short paragraph that concludes the results section by synthesizing the key findings of the study . Highlight the most important findings you want readers to remember as they transition into the discussion section. This is particularly important if, for example, there are many results to report, the findings are complicated or unanticipated, or they are impactful or actionable in some way [i.e., able to be pursued in a feasible way applied to practice].
NOTE: Always use the past tense when referring to your study's findings. Reference to findings should always be described as having already happened because the method used to gather the information has been completed.
III. Problems to Avoid
When writing the results section, avoid doing the following :
- Discussing or interpreting your results . Save this for the discussion section of your paper, although where appropriate, you should compare or contrast specific results to those found in other studies [e.g., "Similar to the work of Smith [1990], one of the findings of this study is the strong correlation between motivation and academic achievement...."].
- Reporting background information or attempting to explain your findings. This should have been done in your introduction section, but don't panic! Often the results of a study point to the need for additional background information or to explain the topic further, so don't think you did something wrong. Writing up research is rarely a linear process. Always revise your introduction as needed.
- Ignoring negative results . A negative result generally refers to a finding that does not support the underlying assumptions of your study. Do not ignore them. Document these findings and then state in your discussion section why you believe a negative result emerged from your study. Note that negative results, and how you handle them, can give you an opportunity to write a more engaging discussion section, therefore, don't be hesitant to highlight them.
- Including raw data or intermediate calculations . Ask your professor if you need to include any raw data generated by your study, such as transcripts from interviews or data files. If raw data is to be included, place it in an appendix or set of appendices that are referred to in the text.
- Be as factual and concise as possible in reporting your findings . Do not use phrases that are vague or non-specific, such as, "appeared to be greater than other variables..." or "demonstrates promising trends that...." Subjective modifiers should be explained in the discussion section of the paper [i.e., why did one variable appear greater? Or, how does the finding demonstrate a promising trend?].
- Presenting the same data or repeating the same information more than once . If you want to highlight a particular finding, it is appropriate to do so in the results section. However, you should emphasize its significance in relation to addressing the research problem in the discussion section. Do not repeat it in your results section because you can do that in the conclusion of your paper.
- Confusing figures with tables . Be sure to properly label any non-textual elements in your paper. Don't call a chart an illustration or a figure a table. If you are not sure, go here .
Annesley, Thomas M. "Show Your Cards: The Results Section and the Poker Game." Clinical Chemistry 56 (July 2010): 1066-1070; Bavdekar, Sandeep B. and Sneha Chandak. "Results: Unraveling the Findings." Journal of the Association of Physicians of India 63 (September 2015): 44-46; Burton, Neil et al. Doing Your Education Research Project . Los Angeles, CA: SAGE, 2008; Caprette, David R. Writing Research Papers. Experimental Biosciences Resources. Rice University; Hancock, Dawson R. and Bob Algozzine. Doing Case Study Research: A Practical Guide for Beginning Researchers . 2nd ed. New York: Teachers College Press, 2011; Introduction to Nursing Research: Reporting Research Findings. Nursing Research: Open Access Nursing Research and Review Articles. (January 4, 2012); Kretchmer, Paul. Twelve Steps to Writing an Effective Results Section. San Francisco Edit ; Ng, K. H. and W. C. Peh. "Writing the Results." Singapore Medical Journal 49 (2008): 967-968; Reporting Research Findings. Wilder Research, in partnership with the Minnesota Department of Human Services. (February 2009); Results. The Structure, Format, Content, and Style of a Journal-Style Scientific Paper. Department of Biology. Bates College; Schafer, Mickey S. Writing the Results. Thesis Writing in the Sciences. Course Syllabus. University of Florida.
Writing Tip
Why Don't I Just Combine the Results Section with the Discussion Section?
It's not unusual to find articles in scholarly social science journals where the author(s) have combined a description of the findings with a discussion about their significance and implications. You could do this. However, if you are inexperienced writing research papers, consider creating two distinct sections for each section in your paper as a way to better organize your thoughts and, by extension, your paper. Think of the results section as the place where you report what your study found; think of the discussion section as the place where you interpret the information and answer the "So What?" question. As you become more skilled writing research papers, you can consider melding the results of your study with a discussion of its implications.
Driscoll, Dana Lynn and Aleksandra Kasztalska. Writing the Experimental Report: Methods, Results, and Discussion. The Writing Lab and The OWL. Purdue University.
- << Previous: Insiderness
- Next: Using Non-Textual Elements >>
- Last Updated: Aug 21, 2024 8:54 AM
- URL: https://libguides.usc.edu/writingguide
Generate accurate APA citations for free
- Knowledge Base
- APA Style 7th edition
- How to write an APA results section
Reporting Research Results in APA Style | Tips & Examples
Published on December 21, 2020 by Pritha Bhandari . Revised on January 17, 2024.
The results section of a quantitative research paper is where you summarize your data and report the findings of any relevant statistical analyses.
The APA manual provides rigorous guidelines for what to report in quantitative research papers in the fields of psychology, education, and other social sciences.
Use these standards to answer your research questions and report your data analyses in a complete and transparent way.
Instantly correct all language mistakes in your text
Upload your document to correct all your mistakes in minutes
Table of contents
What goes in your results section, introduce your data, summarize your data, report statistical results, presenting numbers effectively, what doesn’t belong in your results section, frequently asked questions about results in apa.
In APA style, the results section includes preliminary information about the participants and data, descriptive and inferential statistics, and the results of any exploratory analyses.
Include these in your results section:
- Participant flow and recruitment period. Report the number of participants at every stage of the study, as well as the dates when recruitment took place.
- Missing data . Identify the proportion of data that wasn’t included in your final analysis and state the reasons.
- Any adverse events. Make sure to report any unexpected events or side effects (for clinical studies).
- Descriptive statistics . Summarize the primary and secondary outcomes of the study.
- Inferential statistics , including confidence intervals and effect sizes. Address the primary and secondary research questions by reporting the detailed results of your main analyses.
- Results of subgroup or exploratory analyses, if applicable. Place detailed results in supplementary materials.
Write up the results in the past tense because you’re describing the outcomes of a completed research study.
Scribbr Citation Checker New
The AI-powered Citation Checker helps you avoid common mistakes such as:
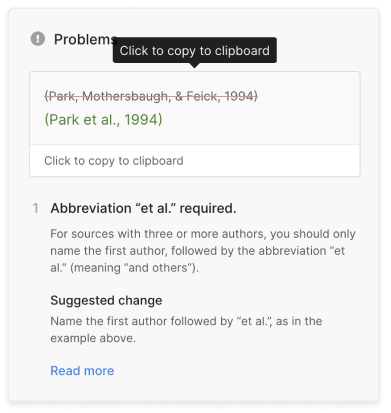
- Missing commas and periods
- Incorrect usage of “et al.”
- Ampersands (&) in narrative citations
- Missing reference entries
Before diving into your research findings, first describe the flow of participants at every stage of your study and whether any data were excluded from the final analysis.
Participant flow and recruitment period
It’s necessary to report any attrition, which is the decline in participants at every sequential stage of a study. That’s because an uneven number of participants across groups sometimes threatens internal validity and makes it difficult to compare groups. Be sure to also state all reasons for attrition.
If your study has multiple stages (e.g., pre-test, intervention, and post-test) and groups (e.g., experimental and control groups), a flow chart is the best way to report the number of participants in each group per stage and reasons for attrition.
Also report the dates for when you recruited participants or performed follow-up sessions.
Missing data
Another key issue is the completeness of your dataset. It’s necessary to report both the amount and reasons for data that was missing or excluded.
Data can become unusable due to equipment malfunctions, improper storage, unexpected events, participant ineligibility, and so on. For each case, state the reason why the data were unusable.
Some data points may be removed from the final analysis because they are outliers—but you must be able to justify how you decided what to exclude.
If you applied any techniques for overcoming or compensating for lost data, report those as well.
Adverse events
For clinical studies, report all events with serious consequences or any side effects that occured.
Descriptive statistics summarize your data for the reader. Present descriptive statistics for each primary, secondary, and subgroup analysis.
Don’t provide formulas or citations for commonly used statistics (e.g., standard deviation) – but do provide them for new or rare equations.
Descriptive statistics
The exact descriptive statistics that you report depends on the types of data in your study. Categorical variables can be reported using proportions, while quantitative data can be reported using means and standard deviations . For a large set of numbers, a table is the most effective presentation format.
Include sample sizes (overall and for each group) as well as appropriate measures of central tendency and variability for the outcomes in your results section. For every point estimate , add a clearly labelled measure of variability as well.
Be sure to note how you combined data to come up with variables of interest. For every variable of interest, explain how you operationalized it.
According to APA journal standards, it’s necessary to report all relevant hypothesis tests performed, estimates of effect sizes, and confidence intervals.
When reporting statistical results, you should first address primary research questions before moving onto secondary research questions and any exploratory or subgroup analyses.
Present the results of tests in the order that you performed them—report the outcomes of main tests before post-hoc tests, for example. Don’t leave out any relevant results, even if they don’t support your hypothesis.
Inferential statistics
For each statistical test performed, first restate the hypothesis , then state whether your hypothesis was supported and provide the outcomes that led you to that conclusion.
Report the following for each hypothesis test:
- the test statistic value,
- the degrees of freedom ,
- the exact p- value (unless it is less than 0.001),
- the magnitude and direction of the effect.
When reporting complex data analyses, such as factor analysis or multivariate analysis, present the models estimated in detail, and state the statistical software used. Make sure to report any violations of statistical assumptions or problems with estimation.
Effect sizes and confidence intervals
For each hypothesis test performed, you should present confidence intervals and estimates of effect sizes .
Confidence intervals are useful for showing the variability around point estimates. They should be included whenever you report population parameter estimates.
Effect sizes indicate how impactful the outcomes of a study are. But since they are estimates, it’s recommended that you also provide confidence intervals of effect sizes.
Subgroup or exploratory analyses
Briefly report the results of any other planned or exploratory analyses you performed. These may include subgroup analyses as well.
Subgroup analyses come with a high chance of false positive results, because performing a large number of comparison or correlation tests increases the chances of finding significant results.
If you find significant results in these analyses, make sure to appropriately report them as exploratory (rather than confirmatory) results to avoid overstating their importance.
While these analyses can be reported in less detail in the main text, you can provide the full analyses in supplementary materials.
To effectively present numbers, use a mix of text, tables , and figures where appropriate:
- To present three or fewer numbers, try a sentence ,
- To present between 4 and 20 numbers, try a table ,
- To present more than 20 numbers, try a figure .
Since these are general guidelines, use your own judgment and feedback from others for effective presentation of numbers.
Tables and figures should be numbered and have titles, along with relevant notes. Make sure to present data only once throughout the paper and refer to any tables and figures in the text.
Formatting statistics and numbers
It’s important to follow capitalization , italicization, and abbreviation rules when referring to statistics in your paper. There are specific format guidelines for reporting statistics in APA , as well as general rules about writing numbers .
If you are unsure of how to present specific symbols, look up the detailed APA guidelines or other papers in your field.
It’s important to provide a complete picture of your data analyses and outcomes in a concise way. For that reason, raw data and any interpretations of your results are not included in the results section.
It’s rarely appropriate to include raw data in your results section. Instead, you should always save the raw data securely and make them available and accessible to any other researchers who request them.
Making scientific research available to others is a key part of academic integrity and open science.
Interpretation or discussion of results
This belongs in your discussion section. Your results section is where you objectively report all relevant findings and leave them open for interpretation by readers.
While you should state whether the findings of statistical tests lend support to your hypotheses, refrain from forming conclusions to your research questions in the results section.
Explanation of how statistics tests work
For the sake of concise writing, you can safely assume that readers of your paper have professional knowledge of how statistical inferences work.
In an APA results section , you should generally report the following:
- Participant flow and recruitment period.
- Missing data and any adverse events.
- Descriptive statistics about your samples.
- Inferential statistics , including confidence intervals and effect sizes.
- Results of any subgroup or exploratory analyses, if applicable.
According to the APA guidelines, you should report enough detail on inferential statistics so that your readers understand your analyses.
- the test statistic value
- the degrees of freedom
- the exact p value (unless it is less than 0.001)
- the magnitude and direction of the effect
You should also present confidence intervals and estimates of effect sizes where relevant.
In APA style, statistics can be presented in the main text or as tables or figures . To decide how to present numbers, you can follow APA guidelines:
- To present three or fewer numbers, try a sentence,
- To present between 4 and 20 numbers, try a table,
- To present more than 20 numbers, try a figure.
Results are usually written in the past tense , because they are describing the outcome of completed actions.
The results chapter or section simply and objectively reports what you found, without speculating on why you found these results. The discussion interprets the meaning of the results, puts them in context, and explains why they matter.
In qualitative research , results and discussion are sometimes combined. But in quantitative research , it’s considered important to separate the objective results from your interpretation of them.
Cite this Scribbr article
If you want to cite this source, you can copy and paste the citation or click the “Cite this Scribbr article” button to automatically add the citation to our free Citation Generator.
Bhandari, P. (2024, January 17). Reporting Research Results in APA Style | Tips & Examples. Scribbr. Retrieved August 21, 2024, from https://www.scribbr.com/apa-style/results-section/
Is this article helpful?

Pritha Bhandari
Other students also liked, how to write an apa methods section, how to format tables and figures in apa style, reporting statistics in apa style | guidelines & examples, "i thought ai proofreading was useless but..".
I've been using Scribbr for years now and I know it's a service that won't disappoint. It does a good job spotting mistakes”

- Langson Library
- Science Library
- Grunigen Medical Library
- Law Library
- Connect From Off-Campus
- Accessibility
- Gateway Study Center

Email this link
Writing a scientific paper.
- Writing a lab report
- INTRODUCTION
Writing a "good" results section
Figures and Captions in Lab Reports
"Results Checklist" from: How to Write a Good Scientific Paper. Chris A. Mack. SPIE. 2018.
Additional tips for results sections.
- LITERATURE CITED
- Bibliography of guides to scientific writing and presenting
- Peer Review
- Presentations
- Lab Report Writing Guides on the Web
This is the core of the paper. Don't start the results sections with methods you left out of the Materials and Methods section. You need to give an overall description of the experiments and present the data you found.
- Factual statements supported by evidence. Short and sweet without excess words
- Present representative data rather than endlessly repetitive data
- Discuss variables only if they had an effect (positive or negative)
- Use meaningful statistics
- Avoid redundancy. If it is in the tables or captions you may not need to repeat it
A short article by Dr. Brett Couch and Dr. Deena Wassenberg, Biology Program, University of Minnesota
- Present the results of the paper, in logical order, using tables and graphs as necessary.
- Explain the results and show how they help to answer the research questions posed in the Introduction. Evidence does not explain itself; the results must be presented and then explained.
- Avoid: presenting results that are never discussed; presenting results in chronological order rather than logical order; ignoring results that do not support the conclusions;
- Number tables and figures separately beginning with 1 (i.e. Table 1, Table 2, Figure 1, etc.).
- Do not attempt to evaluate the results in this section. Report only what you found; hold all discussion of the significance of the results for the Discussion section.
- It is not necessary to describe every step of your statistical analyses. Scientists understand all about null hypotheses, rejection rules, and so forth and do not need to be reminded of them. Just say something like, "Honeybees did not use the flowers in proportion to their availability (X2 = 7.9, p<0.05, d.f.= 4, chi-square test)." Likewise, cite tables and figures without describing in detail how the data were manipulated. Explanations of this sort should appear in a legend or caption written on the same page as the figure or table.
- You must refer in the text to each figure or table you include in your paper.
- Tables generally should report summary-level data, such as means ± standard deviations, rather than all your raw data. A long list of all your individual observations will mean much less than a few concise, easy-to-read tables or figures that bring out the main findings of your study.
- Only use a figure (graph) when the data lend themselves to a good visual representation. Avoid using figures that show too many variables or trends at once, because they can be hard to understand.
From: https://writingcenter.gmu.edu/guides/imrad-results-discussion
- << Previous: METHODS
- Next: DISCUSSION >>
- Last Updated: Aug 4, 2023 9:33 AM
- URL: https://guides.lib.uci.edu/scientificwriting
Off-campus? Please use the Software VPN and choose the group UCIFull to access licensed content. For more information, please Click here
Software VPN is not available for guests, so they may not have access to some content when connecting from off-campus.
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- Int J Endocrinol Metab
- v.17(2); 2019 Apr

The Principles of Biomedical Scientific Writing: Results
Zahra bahadoran.
1 Nutrition and Endocrine Research Center, Research Institute for Endocrine Sciences, Shahid Beheshti University of Medical Sciences, Tehran, Iran
Parvin Mirmiran
2 Department of Clinical Nutrition and Diet Therapy, Faculty of Nutrition Sciences and Food Technology, National Nutrition and Food Technology Research Institute, Shahid Beheshti University of Medical Sciences, Tehran, Iran
Azita Zadeh-Vakili
3 Cellular and Molecular Endocrine Research Center, Research Institute for Endocrine Sciences, Shahid Beheshti University of Medical Sciences, Tehran, Iran
Farhad Hosseinpanah
4 Obesity Research Center, Research Institute for Endocrine Sciences, Shahid Beheshti University of Medical Sciences, Tehran, Iran
Asghar Ghasemi
5 Endocrine Physiology Research Center, Research Institute for Endocrine Sciences, Shahid Beheshti University of Medical Sciences, Tehran, Iran
The “results section” of a scientific paper provides the results related to all measurements and outcomes that have been posted earlier in the materials and methods section. This section consists of text, figures, and tables presenting detailed data and facts without interpretation and discussion. Results may be presented in chronological order, general to specific order, most to least important order, or may be organized according to the topic/study groups or experiment/measured parameters. The primary content of this section includes the most relevant results that correspond to the central question stated in the introduction section, whether they support the hypothesis or not. Findings related to secondary outcomes and subgroup analyses may be reported in this section. All results should be presented in a clear, concise, and sensible manner. In this review, we discuss the function, content, and organization of the “results section,” as well as the principles and the most common tips for the writing of this section.
The “results section” is the heart of the paper, around which the other sections are organized ( 1 ). Research is about results and the reader comes to the paper to discover the results ( 2 ). In this section, authors contribute to the development of scientific literature by providing novel, hitherto unknown knowledge ( 3 ). In addition to the results, this section contains data and statistical information for supporting or refuting the hypothesis proposed in the introduction ( 4 ).
“Results section” should provide an objective description of the main findings, clearly and concisely, without interpretation ( 5 , 6 ). The authors need to use an interesting combination of text, tables, and figures to answer the study questions and to tell the story without diversions ( 7 ). The systemic assessment of published articles highlights the fact that the literature frequently suffers from selective reporting of results only for certain assessed outcomes, selective reporting of statistical analyses, and confused, ambiguous, incomplete, or misleading presentation of data ( 8 , 9 ).
In this section of our series on the principles of biomedical scientific writing ( 10 , 11 ), we describe the function, content, and organization of the “results section” in a scientific paper (mostly for hypothesis-testing papers) and provide common recommendations that can help authors to write this section more effectively.
2. The Function of the Results Section
The function of the “results section” is to present the main results of experiments described in the materials and methods section ( 12 , 13 ) and to present the supporting data in the form of text, tables, and figures ( 13 ). This section should answer the basic question: “What did the authors find in research?” By providing the results, authors try to elucidate the research data, making it to the point and meaningful ( 13 ).
3. Content of the Results Section
The “results section” includes both results and data that are presented in text, tables, and figures. Results are presented in the text; data (the most important) are presented in figures and tables, with a limited amount presented in the text ( 13 ). Statistically relevant parameters including sample size, P values, and the type of statistics used are also presented in this section ( 13 ).
3.1. Difference Between Data and Results
Data and results are not the same ( 14 ); providing results but no data vs. data but no results should be avoided ( 14 , 15 ). Results are general statements in the main text that summarize or explain what the data (facts and numbers) show ( 13 , 14 ); in other words, results are text descriptions of what is important about data ( 16 ) and give meaning to the data ( 15 ). When reporting data or results, make sure that they are logical ( 2 ). See Box 1 for more differences between results and data.
| Data | Results |
|---|---|
| Are the facts (often numbers) obtained from experiments or observations. | Are the meaning and interpretation of data |
| Can be presented as raw (e.g. concentration of a measured variable), summarized (e.g. mean and SD), or transformed (e.g. percentage). | Are expressed as statements that explain or summarize what the data show |
| Can rarely stand alone | May have a direction (e.g. decrease, increase) or magnitude, e.g. 2-fold, 10% increased |
| May contain statistical significance, e.g. value | |
| E.g. mean (SD) fasting blood glucose was 180 (20) mg/dL in patients with type 2 diabetes. Mean fasting blood glucose was 95 (5) mg/dL in non-diabetic subjects. | E.g. mean fasting blood glucose was significantly higher in patients with type 2 diabetes than in non-diabetic subjects [180 (20) vs. 95 (5) mg/dL, = 0.010] . |
a The text presented in square brackets is data and the remainder is a result.
3.2. The Appropriate Format for Presenting Data/Results
Depending on how the data best support the findings of the study, the “results section” is structured as text, tables, and figures ( 12 ) and should consist of a dynamic interplay between text and figures/tables; the most important data are usually presented in both formats ( 17 ). The reader should select the mode of presentation in a way that optimizes comprehension of the data; however, as a general rule, if you want to present three or fewer numbers, you should use a sentence; otherwise, you consider a table or a graph ( 18 ).
Selecting the best format for presenting results/data depends on the level of details (exact values or patterns) to present ( 19 ). Tables are useful to present specific information or exact values ( 19 ), and function as reference tools for readers ( 20 ) whereas figures are useful to show comparisons and patterns ( 19 ), functioning as analytic tools ( 20 ).
Tables are meant to summarize large amounts of data, to organize and display data more clearly than words, to compare groups of data, to simplify found information, and to facilitate calculations ( 19 ). A table typically has three or more interrelated columns and three or more interrelated rows; otherwise, presenting the information in the text may be more appropriate ( 19 ).
The functions of figures include: (1) showing the underlying patterns of data that are not presentable in text or tables, (2) displaying data more clearly than they can be done in text or tables, (3) more summarizing a large amount of data than they can be done in text or tables, and (4) improving the understanding and locating the specific information easily and rapidly ( 21 ).
3.3. Results
The primary content of this section includes the most relevant (but not all) results corresponding to the central question posed in the introduction section, whether they support the hypothesis or not ( 12 , 13 ). The secondary findings, e.g., results related to secondary outcomes and subgroup analyses, may also be reported in this section ( 22 ). Results must be presented for both experimental and control groups ( 13 ). Results of each item mentioned in the materials and methods should be given in the results section ( 12 , 15 ).
The text of the “results section” should state and summarize the main results and explain the data presented within tables and/or figures ( 23 ); reiteration of all numbers presented in tables and figures is not recommended ( 22 ); however, readers must be given the main messages derived from a table or figure without having to interpret the data themselves ( 7 ). It means that if there is a large amount of data in a table or figure, restating a key piece of data in the text is acceptable and helps the reader zero in on important data ( 14 ).
3.3.1. Reporting Negative Findings
Authors are highly recommended excluding irrelevant results but not ignoring valid anomalous results that contradict the research hypothesis or do not support the current scientific literature ( 22 ). The Feynman, says “if you are doing an experiment, you should report everything that you think might make it invalid-not only what you think is right about it” ( 24 ). Although reporting null or negative findings is not as straightforward as positive findings, it may lead to reexamining current scientific thinking, and guide scientists towards unabridged science ( 25 ). Reporting negative findings can also prevent the replication of the study and prevent the waste of time and resources ( 25 ). The ignorance of null or negative findings also leads to an overestimation of an effect size or treatment effect in available data ( 9 ).
3.3.2. Referring to Unpublished Results
Referring to unpublished results is not recommend unless there is a strong argument supporting their inclusion ( 14 ); therefore, authors are advised to avoid using the term “data not shown” ( 4 ).
3.3.3. Methods or Interpretation in the Results Section
Generally, the “results section” is not the place for presenting methods and experimental details or interpreting data ( 14 ). When experiments are described in this section, if a result leads to additional experiments, it is better to report the new experimental details in the “results section” ( 14 ). Sometimes authors want to refer to a specific experiment or method in results; in these cases, they should not repeat experimental details, but preferably use a transition phrase to link methods with results ( 14 ). To justify the rationale behind the experiment, using topic sentences/phrases (e.g. in order to determine whether…) provides an overview before giving details ( 12 ); however, in this case, the method statement should not be used as a topic sentence and the main verbs should describe results, not methods (e.g., “ when propranolol was administered during normal ventilation, phospholipids decreased ”; here “ method ” is subordinated in a transition clause and result is the main clause) ( 13 ). Two patterns of sentence structure are recommended for including methods in a result statement: making the method the subject of the sentence or stating the method using a transition phrase or clause and the result in the main clause ( 13 ).
The traditional view of writing the “results section” is just to report data and results without any interpretation; accordingly, the result is not expected to contain statements that need to be referenced (comparisons of findings) ( 13 , 26 ). In another view, some interpretation or brief comparisons that do not fit into the discussion may be included ( 13 , 27 ).
Data are facts and numbers, mostly presented as non-textual elements (usually in tables and figures) where they are easy to read ( 13 , 14 , 28 ). A limited amount of data may also be presented in the text, following a result statement ( 13 ) although too much data in the text make it too long ( Box 1 ) ( 28 ). Data may be in the form of raw data, summarized data, or transformed data ( 13 ); however, it is suggested that raw data (i.e. patients’ records, individual observations) not be presented in results ( 12 ). Note that numerical data are absolute while some data, e.g. microscopic data, are subjective ( 2 ).
3.4.1. Non-Textual Elements
Providing study findings visually, rather than entire textualizing, enables authors to summarize a great deal of data compactly within the text with an appropriate reference; some images convey more than words ( 29 ). The primary purpose of non-textual elements, i.e. tables, graphs, figures, and maps, is to present data such that they can be easily and quickly grasped ( 23 ) while being more informative than when appearing in the text ( 6 ). Tables and figures should be complete/comprehensible, being able to stand alone without the text ( 5 , 12 ).
Non-textual elements should be referred to in the text at the appropriate point ( 5 , 6 , 12 ). Location statements, i.e. statements referring to non-textual elements, may be presented in different patterns (e.g., A. X is shown in table/figure; B. table/figure shows; C. see table/figure; D. as shown in table/figure); pattern B is more and pattern C is less common ( 27 ).

Some general tips about using non-textual elements in the “results section” are reviewed in Box 2 . The most common rules in organizing tables and figures are given in the following. For more information about designing different types of tables/figures/graphs, please refer to additional references ( 7 , 19 , 20 , 30 , 31 ).
| Tips |
|---|
| Give a caption to each element consisting of a number and a title |
| Avoid using abbreviations in the title of tables or the legend of figures |
| Keep the table title and figure legend brief but sufficiently detailed to explain the data included |
| Do not overload the title with details |
| Put the elements within the text, or include them in the rest of the manuscript; do not use both approaches |
| Distinguish the element from any appendix materials provided at the end of the manuscript (if placed at the end) |
| Put each element as close as possible to where it is first mentioned in the text (if placed within the text) |
| Use an explicit number for each table, figure, etc. |
| Refer to each element appropriately within the text and if needed explain it |
| Use parentheses when referring to elements within the text |
| Have a consistent appearance for the elements, e.g. use a uniform box or frame and a uniform font |
| Use footnotes or captions to explain any unclear data |
3.4.1.1. Tables
The use of tables is an effective way to summarize demographic information and descriptive statistics ( 23 ). Note that tables must have a purpose and be integrated into the text ( 21 ). Tables are most useful to present counts, proportions, and percentages ( 8 ), and are appropriate also for presenting details especially when exact values matter ( 32 ), being are more informative than graphs ( 29 ). However, limited information should be presented in tables; otherwise, most readers find them difficult to read and thus, may ignore them ( 5 , 23 ). Data in tables can be arranged horizontally or vertically; whenever possible, primary comparisons are preferably presented horizontally from left to right ( 19 ).
3.4.1.1.1. Basic Elements of Tables
Tables usually have at least six elements: (1) table number, (2) table title, (3) row headings (stubs), and (4) column headings (boxes), identifying information in rows and columns, (5) data in data field, and (6) horizontal lines (rules). Most also have footnotes, row subheadings, spanner headings (identifying subgroups in column headings), and expanded forms of abbreviations in the table ( 19 , 21 , 31 , 33 ).
The table title should clearly state what appears in it and provide sufficient information on the study, i.e. provide a context helping readers interpret the table information ( 19 ). Some specific details may also be provided including the type and number of subjects or the period of study ( 30 ). For developing the title of a table, one can describe the main cell entries, followed by qualification or more description ( 32 ). The table’s title is presented as a phrase not a full sentence ( 19 ). Authors need to refer to the journal’s style for rules on which words in titles are capitalized.
As a rule, comparing two (or even three) numbers should be side-by-side rather than above and below ( 30 ). Column and row headings help readers find information and they should be included group sizes and measurement units ( 19 ). Tables should be in borderless grids of rows and columns ( 5 , 32 ) with no vertical rule and limited horizontal rules ( 32 ). The first column of a table includes usually a list of variables that are presented in the table; although the first column usually does not need a header, sometimes a simple description of what appears in each row may be provided as the heading of the first column. Units for variables may be placed in parentheses immediately below the row descriptions ( 30 ).
Headings for other columns should also be informative without vague labels, e.g. group A, group B, group C, etc.; instead, a brief description summarizing group characteristics is used ( 30 ). The last column may show P values for comparison between study groups ( 34 ), except for randomized clinical trials, where P values are not needed to compare baseline characteristics of participants ( 7 ). The first letters of lines and column headings in tables should be capitalized.
The fields of tables are points at which columns and rows intersect ( 19 ). Cells of a table are the data field of the table, other than those containing row and column headings ( 21 ). Cells contain information as numerals, text, or symbols ( 19 ). Every cell must contain information; if no information is available, one can use NA in the cell and define it in the footnote as not available or not applicable; alternatively, a dash mark may be inserted ( 19 ). The content of columns need to be aligned ( 19 ); words are usually left aligned, numerals are aligned at decimals, parenthesis, and factors of 10 ( 19 , 21 ).
Table footnotes should be brief, and define abbreviations, provide statistical results, and explain discrepancies in data, e.g., “percentages do not total 100 because of rounding” ( 19 , 30 ). In addition to asterisks usually used to show statistical significance ( 33 ), the following symbols are used, in sequence, for further notes: †, ‡, §, ¶, #, ††, ‡‡ ( 30 ).
3.4.1.1.2. Different Types of Tables
Table of lists, table of baseline or clinical characteristics of subjects, table of comparisons, and table of multivariable results are various types of tables that may be used ( 30 ). The table’s format should be selected according to the purpose of the table ( 30 ). A table of lists just presents a list of items including diagnostic criteria or causes of a disease; it is critical to arrange such tables based on their contents by order (e.g., alphabetical order) or their importance (most to least) ( 30 ). Tables of study participants’ characteristics usually provide a general overview of the essential characteristics of subjects, such as age, sex, race, disease stage, and selected risk factors ( 30 ). The table of comparisons (≥ two groups) provides details for each group and differences between the groups. Tables of multivariable results elaborate results of statistical analyses assessing relationships between predictor (independent) and outcome (dependent) variables, and usually include regression coefficients, standard errors, slopes, partial correlation coefficients, and P values or odds ratio, hazard ratios, and 95% confidence intervals for regression models ( 30 ).
3.4.1.2. Figures
Graphical elements convey the important messages of research ( 20 ). A figure is “any graphical display to present information or data” ( 20 ), and it effectively presents complicated patterns ( 32 ), best used for presenting an important point at a glance or indicating trends or relationships ( 20 ). Like tables, figures should have a purpose and be integrated with the rest of the text ( 21 ).
3.4.1.2.1. Basic Elements of Figures
Most figures that present quantitative information (charts and graphs) have at least seven elements, including figure number, figure caption/legend, data field, vertical scale, horizontal scale, labels, and data (plotting symbols, lines, and so on) ( 21 ). Some figures also have reference lines in the data field to help orient readers and keys that identify data ( 21 ).
Figure caption/legend, usually given below the figure, describes the figure and must reflect the figure entirely, independent of the main text ( 21 , 31 ). For the figure to stand alone, a figure legend needs to be included four parts (a brief title, experimental or statistical information/details, definitions of symbols, line, or bar patterns, and abbreviations) ( 31 ).
Data field is a space in the figure in which data are presented; it is usually bordered on the left by the X-axis (abscissa) and on the bottom by the Y-axis (ordinate) ( 20 , 21 ). Labels identify the variables graphed and the units of measurement ( 21 ). Figure lines should be broad and the labeling text should be large enough to be legible after reduction to a single- or two-column size ( 32 ). Appropriate font size should be used to maintain legibility after fitting figures to publication size ( 31 ).
Scales on each axis should match the data range and be slightly above the highest value ( 20 ). Symbols should be uniform across the figures ( 20 ). The data point symbols should be easily distinguishable; using black and white circles (● - ∘) is the easiest way when two are needed ( 31 ); if more are needed, using up-pointing triangles (▲ - Δ) and squares (■ - □) is suggested ( 31 ). Using symbols, line types, and colors is also effective in differentiating important strata in figures ( 8 ).
3.4.1.2.2. Emphasizing Important Data on Figures
To make figures visually efficient, the subordination of all non-data elements vs. data elements is advised (gridlines should be used as thin as possible and very faint). Directly labeling objects, instead of legends, may keep readers’ attention on the most important parts of the figure ( 8 ). Using different line weights may also be helpful to emphasize the important information/data in figures ( 31 ). The use of color, shading, or 3D perspectives is not suggested unless they serve a specific explanatory function in figure ( 8 ).
3.4.1.2.3. Different Types of Figures
Two major categories of figures are statistical figures (graphs) and non-statistical figures (clinical images, photographs, diagrams, illustrations, and textual figures) ( 20 ). Graphs are suitable for presenting relationships whereas non-statistical figures are used to confirm findings or provide explanatory information ( 20 ).
In statistical figures, selecting a graphical format (bar graph, line graph, dot plot, and scatterplot) is done according to the type of relationship that authors wish to communicate ( 20 ); for example, line graphs are appropriate for showing trends and bar graphs for magnitudes ( 20 ). Using a graphing format that is easy to interpret is preferred ( 20 ); pie graphs are sparingly used because comparing different angles is complicated with them ( 20 ). Graphs should accurately represent findings; when possible, scales should start at zero, and figure axes should not be altered in order to make data more meaningful ( 20 ).
Non-statistical figures are those that visually present information that does not contain data ( 20 ). Clinical images and photographs [ultrasonograms, computed tomographic scans (CT scans), magnetic resonance images (MRI), images of patients, tissue samples, microscopic findings, and so on] provide absolute proof of findings ( 20 ). Illustrations are used for explaining structures (parts of a cell), mechanisms, and relationships ( 20 ). Diagrams (flowcharts, algorithms, pedigrees, and maps) are useful for displaying complex relations ( 20 ). Textual figures, containing only text, are mostly used for describing steps of a procedure or summarizing guidelines ( 20 ). For photographs, patient information or identifiers should be removed ( 20 ).
3.5. Statistics in the Results Section
Statistics in the “results section” must report data in a way that enables readers to assess the degree of experimental variation and to estimate the variability or precision of the findings ( 22 ). For more details, one can see SAMPL (Statistical Analysis and methods in the Published Literature) guidelines ( 35 ). To report normally distributed data, the mean and estimated variation from mean should be stated ( 13 ). Variability should be reported using standard deviation (SD), which is a descriptive statistic ( 36 ) and reflects the dispersion of individual sample observation of the sample mean ( 37 ). The standard error (SE), an inferential statistic ( 36 ) reflecting the theoretical dispersion of sample means about some population means, characterizes uncertainty about true values of population means ( 37 ). It is useful for assessing the precision of an estimator ( 36 ) and is not an appropriate estimate of the variability in observations ( 37 ). Using “mean (SD or SE)” is preferred to “mean ± SD or SE” because the “±” sign can cause confusion ( 22 ). Increasing sample size decreases SE but not SD ( 36 ). To report data with a skewed distribution, the median and the interquartile range (between 25th and 75th percentiles) should be provided ( 22 ).
To report risk, rates, and ratios, one should use a type of rate (incidence rate, survival rate), ratio (odds ratio, hazards ratio), or risk (absolute risk, relative risk, relative risk reduction) ( 35 ). The measure of precision (95% CI) for estimated risks, rates, and ratios should also be provided ( 35 ). For correlation analysis, the exact values of the correlation coefficient and 95% CI should be reported. Describing correlation using qualitative words (low, moderate, high) without providing a clear definition is not acceptable ( 35 ). Results of regression analysis should include regression coefficients (β) of each explanatory variable, corresponding 95% CI and/or P value and a measure of the “goodness-of-fit” of the model ( 35 ).
3.5.1. Significance Levels
A P value is the probability of consistency between data and the hypothesis being tested ( 38 ). Reporting the exact P values ( P = 0.34 or P = 0.02) rather than the conventional P ( P < 0.05) is recommended for all primary analyses ( 12 , 37 ) as it conveys more information ( 37 ). The use of the term “partially significant” or “marginally significant”, where the P value is almost significant (e.g. P = 0.057) is not acceptable if the significance level is defined as P = 0.05 ( 39 ). Some, however, argue that it is not always necessary to stick to P = 0.05 for the interpretation of results and it is better to report the exact P value and confidence interval for the estimator ( 40 ).
The use of the 95% confidence interval (95% CI) can provide further information compared to P values per se, and prefigures the direction of the effect size (negative or positive), its magnitude, and the degree of precision ( 17 ). A confidence interval characterizes uncertainty about the true value of population parameters ( 37 ). It is essential to provide the sample size (n) and probability values for tests of statistical significance ( 13 ).
Statements about significance must be qualified numerically ( 41 ). In the text, it is suggested that P values be reported as equalities rather than as inequalities in relation to the alpha criterion ( 41 ). In tables and figures, inequalities may be useful for groups of data ( 41 ) where asterisks *, **, and *** are usually used to show statistical significance at 0.05, 0.01, and 0.001 probability levels, respectively ( 33 ).
Although not consistent, P values < 0.001 are reported as P < 0.001; for 0.001 ≤ P values < 0.01, a three-significant digit is recommended, e.g. P = 0.003; for 0.01 ≤ P values < 0.1, a two-significant digit is sufficient (e.g. P = 0.05); for 0.1 ≤ P values ≤ 0.9, a one-significant digit is sufficient (e.g. P = 0.4); and P values > 0.9 are reported as P > 0.9 ( 42 ). For genome-wide association studies, the power of 10 is used for reporting P values, e.g. 6 × 10 -9 ( 42 ). It is generally suggested that zero be used before a decimal point when the value is below one, e.g. 0.37 ( 43 ). According to the American Psychological Association, zero before a decimal point is used for numbers that are below one, but it can also be used for values that may exceed one (e.g. 0.23 cm). Therefore, when statistics cannot be greater than one (e.g. correlations, proportions, and P values), do not use a zero before decimal fraction, e.g. P = .028 not P = 0.028 ( 18 ); this recommendation, however, is not always adopted by everyone. The international standard is P (large italic) although both ‘p’ and ‘P’ are allowed ( 40 ).
4. Organization of the Results Section
There are different ways for organizing the “results section” including ( 1 , 12 , 14 , 22 , 44 ): (1) chronological order, (2) general to specific, (3) most to least important, and (4) grouping results by topic/study groups or experiment/measured parameters. Authors decide which format is more appropriate for the presentation of their data ( 12 ); anyway, results should be presented in a logical manner ( 4 ).
4.1. Different Ways of Organizing the Results Section
4.1.1. chronological order.
The best order for organizing “results section” may be the chronological order ( 22 ). It is considered as the most straightforward approach using subheadings that parallel methods ( 14 ). This order facilitates referring to a method associated with a given result ( 14 ) such that results are presented in the same order as methods ( 15 ).
4.1.2. General to Specific
This format is mostly used in clinical studies involving multiple groups of individuals receiving different treatments ( 14 ). The “results section” usually proceeds from general to more specific findings ( 1 ). Characteristics of the overall study population (sex and age distribution and dropouts) are first given ( 14 ), followed by data and results for each group starting with the control group or the group receiving the standard treatment ( 14 ); finally, the disease group or group receiving the experimental treatment are addressed ( 14 ). As a general rule, secondary results should be given after presenting more important (primary) results, followed by any supporting information ( 22 ). A common order is stating recruitment/response, characteristics of the sample/study participants, findings from the primary analyses, findings from secondary analyses, and any additional or unexpected findings ( 17 ). In other words, the “results section” should be initiated by univariate statistics, followed by bivariate analyses to describe associations between explanatory and outcome variables; finally, it gets through by any multivariate analyses ( 7 ).
4.1.3. Most to Least Important
This format is used in case that the order of presenting results is not critical to their being comprehendible and allows the author to immediately highlight important findings ( 14 ). Results that answer the main question are presented at the beginning of the “results section,” followed by other results in next paragraphs ( 13 ).
4.1.4. Grouping by Topic or Experiment
Comparison of the diagnostic and analytical performance of a number of assays for analytes is an example of using this format ( 14 ).
4.2. Paragraphing of the Results Section
The “results section” may be initiated by two approaches: (1) by giving a general (not detailed) overview of the experiment and (2) by going directly to the results by referring to tables or figures ( 44 ). The first paragraph of this section, along with table 1, describes the characteristics of the study population (number, sex, age, and symptoms) ( 23 ). These data show the comparability of the study groups at baseline and the distribution of potential confounders between groups, as a source of bias that can affect the study findings ( 7 ). It allows the reader to decide whether or not the case and control groups are similar and represent the patient population in their private practice ( 23 ).
For clinical trials, the number of patients completing the protocol in each treatment/study group, the number of patients lost to follow-up, and the number and reasons for excluded/withdrawn subjects should be given. Commenting on whether baseline characteristics of study groups are statistically similar or different is also important ( 1 ). For further information, authors can consult reporting guidelines for the main study types available at http://www.equator-network.org.
The number of the middle paragraphs depends on the number of research questions/hypotheses and the types of statistical analyses; each hypothesis or specific analysis typically devotes at least a paragraph to itself ( 1 ). Figure legends, description of the methods and results for control groups should not be given at the beginning of paragraphs, as they do not narrate the story ( 28 ). However, sometimes, it is needed that results of the control group are presented first (e.g. for establishing the stability of baseline) ( 13 ).
5. Emphasizing Important Results
Since not all results are equally important, the reader must be able to distinguish important results and authors have to emphasize important information and de-emphasize less important information ( 13 ). There are various techniques for emphasizing important information, including condensing or omitting less important information, subordinating less important information, placing important results at the power position, and labeling, stating, and repeating important information ( 13 ).
For condensing or omitting less important information, you should be careful not to duplicate/repeat data in tables and figures or repeat them in the text ( 4 , 6 , 12 ); one or two values from tables/figures can be repeated in the text for emphasis ( 13 ).
For subordinating less important information, one should not use table titles, figure legends or methods statement as a topic sentence in the text ( 13 , 22 ). Instead, after stating the first result relevant to the table/figure, you can cite it in parenthesis ( 13 ). Since a result states a message and creates an expectation, it is a more powerful topic sentence than a figure legend or table title ( 13 ). Sometimes, control results can be subordinated by incorporating them into experimental results ( 13 ).
To highlight more important results (those that help answer questions), authors can put these results at the beginning of paragraphs, the strongest power position ( 12 , 22 , 28 ), followed by supporting details and control results ( 28 ).
Moreover, key findings may receive more attention by using a signal (e.g. we found or we observed) at the beginning of the sentence ( 13 ).
6. Other Considerations
6.1. length and paragraphing.
To see the forest for the tree, the “results section” should be as brief and uncluttered as possible ( 13 ), which can be accomplished by having a well-organized “materials and methods” section ( 3 ) and avoiding unnecessary repetition ( 13 ); for example, similar results for several variables can be reported together. The “results section” of an original manuscript usually includes 2 - 3 pages (~1000 words) with a 1.5 line spacing, font size 11 (including tables and figures) ( 45 ), and 4 - 9 paragraphs (each 130 words) on average ( 45 ); a paragraph should be devoted to one or more closely related figures ( 4 ).
Presenting additional results/data as supplementary materials is a suggestion for keeping the “results section” brief ( 17 ). In addition to save the text space, supplementary materials improve the presentation and facilitate communications among scientists ( 46 , 47 ). According to Springer, supplementary materials can be used for presenting data that are not needed to support the major conclusions but are still interesting. However, keep in mind that the unregulated use of supplementary materials is harmful to science ( 47 ). Supplementary materials should be referred to at the appropriate points in the main text.
For referring to results obtained in hypothesis testing studies, using past tenses is recommended ( 4 , 12 - 14 ); non-textual elements should be referred using present tenses, e.g. “as seen in table 1 …” or “table 1 shows …” in descriptive studies, results are reported in the present tense ( 13 ).
6.3. Word Choice
Although adverbs/adjectives are commonly used to highlight the importance of results, it is recommended altogether avoiding the use of such qualitative/emotive words in the “results section” ( 7 , 13 ). Some believe that qualitative words should not be used because they may imply an interpretation of findings ( 17 ). In biomedical publications, the terms ‘significant, significance, and significantly’ (followed by P values) are used to show statistical relationships and should not be used for other purposes for which, other terms such as substantial, considerable, or noteworthy can be used ( 14 ). See Box 3 for appropriate word choice for the “results section.”
| Do's |
|---|
| Use straightforward verbs for stating results, e.g. show, indicate, demonstrate, highlight, identify, detect, observe, find, and confirm |
| Use “significant” or “significantly” just for statistical significance |
| Be careful about using negative sentences: |
| Instead of using double negatives, be straightforward and use positive terms |
| Make the sentence clear by omitting negative words or negative sentence constructions, e.g. “There was no significant interaction…” instead of “We did not find a statistical interaction ….” |
| Do not use “reveal” to state the results because it is a funny word that suggests something was found perhaps by magic. |
| Do not use emotive words to describe the significance of the results, e.g. interestingly, unfortunately, curiously, remarkably, inexplicably, importantly, crucially, and critically. |
| Do not use the word “level” instead of “concentration.” |
In the “results section,” to make a comparison between the results, i.e. stating the similarity/equivalence or difference/non-equivalence, using appropriate signals is recommended ( 27 ). To show a similarity, a signal to the reader may be used such as “like”, “alike”, “similar to”, and “the same as”; to show differences, the following signals can be used: “but”, “while”, “however”, “in contrast”, “more likely than”, and “less likely than” ( 27 ).
6.4. Reporting Numbers
Numbers play an important role in scientific communication and there are some golden rules for reporting numbers in a scientific paper ( 43 , 48 ). Significant figures (significant digits) should reflect the degree of precision of the original measurement ( 12 ). The number of digits reported for a quantity should be consistent with scientific relevance ( 37 ); for example, a resolution to 0.001 units is necessary for pH but a resolution of < 1 mm Hg is unimportant for blood pressure ( 37 ). Avoid using “about” or “approximately” to qualify a measurement or calculation ( 12 ). The use of percentage for sample sizes of < 20 and decimal for sample sizes of < 100 is not recommended ( 43 ).
The numbers should be spelled out at the beginning of a sentence or when they are less than 10, e.g., twelve students improved… ( 43 ). In a sentence, the authors should be consistent where they use numbers as numerals or spelled-out ( 43 ). Before a unit of a measure, time, dates, and points, numbers should be used as numerals, e.g. 12 cm; 1 h 34 min; at 12:30 A.M., and on a 7-point scale ( 18 ).
A space between the numeral and the unit should be considered, except in the case of %. Because the terms “billion,” “trillion,” and “quadrillion” imply different numbers in Europe and the USA, they should not be used ( 48 ). To express ranges in text, the terms “to” or “through” are preferred to dashes; in tables, the use of dashes or hyphens is recommended ( 48 ).
7. Conclusions
The “results section” of a biomedical manuscript should clearly present findings of the study using an effective combination of results and data. Some dos and don’ts of writing the “results section” are provided in Box 4 . Authors should try to find the best format using a dynamic interplay between text and figures/tables. Results can be organized in different ways including chronological order or most to least important; however, results should be presented in a manner that makes sense.
| Do's |
|---|
| Present demographics or simple descriptive statistics first |
| Describe results from the most to the least important and from the primary outcomes to the secondary outcomes |
| Organize the results section using separate headings as in methods or by categories |
| Make up the results section using a combination of text, tables, and figures |
| Quantify results using appropriate indicators of centrality, probability, and statistical significance values |
| Match each result by its corresponding assessment/measurement method |
| Be focused on results related to the research hypothesis/question |
| Provide units according to the journal style and in a constant manner throughout the text |
| Report all analyses including those unrelated to the main study hypothesis/question |
| Compare the study results with those of previous reports |
| Discuss and interpret the results |
| Restate similar results in both textual and non-textual elements |
| Present raw data |
| Present data lacking units of measurements |
| Present crowded and confusing tables or figures |
Acknowledgments
The authors wish to acknowledge Ms. Niloofar Shiva for critical editing of English grammar and syntax of the manuscript.
Conflict of Interests: It is not declared by the authors.
Funding/Support: Research Institute for Endocrine Sciences supported the study.
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
- My Bibliography
- Collections
- Citation manager
Save citation to file
Email citation, add to collections.
- Create a new collection
- Add to an existing collection
Add to My Bibliography
Your saved search, create a file for external citation management software, your rss feed.
- Search in PubMed
- Search in NLM Catalog
- Add to Search
How to Write an Effective Results Section
Affiliation.
- 1 Rothman Orthopaedics Institute, Philadelphia, PA.
- PMID: 31145152
- DOI: 10.1097/BSD.0000000000000845
Developing a well-written research paper is an important step in completing a scientific study. This paper is where the principle investigator and co-authors report the purpose, methods, findings, and conclusions of the study. A key element of writing a research paper is to clearly and objectively report the study's findings in the Results section. The Results section is where the authors inform the readers about the findings from the statistical analysis of the data collected to operationalize the study hypothesis, optimally adding novel information to the collective knowledge on the subject matter. By utilizing clear, concise, and well-organized writing techniques and visual aids in the reporting of the data, the author is able to construct a case for the research question at hand even without interpreting the data.
PubMed Disclaimer
Similar articles
- Rules to be adopted for publishing a scientific paper. Picardi N. Picardi N. Ann Ital Chir. 2016;87:1-3. Ann Ital Chir. 2016. PMID: 28474609
- How to Write Effective Discussion and Conclusion Sections. Makar G, Foltz C, Lendner M, Vaccaro AR. Makar G, et al. Clin Spine Surg. 2018 Oct;31(8):345-346. doi: 10.1097/BSD.0000000000000687. Clin Spine Surg. 2018. PMID: 29979216
- The rites of writing papers: steps to successful publishing for psychiatrists. Brakoulias V, Macfarlane MD, Looi JC. Brakoulias V, et al. Australas Psychiatry. 2015 Feb;23(1):32-6. doi: 10.1177/1039856214560180. Epub 2014 Dec 2. Australas Psychiatry. 2015. PMID: 25469001
- How to write a research paper. Alexandrov AV. Alexandrov AV. Cerebrovasc Dis. 2004;18(2):135-8. doi: 10.1159/000079266. Epub 2004 Jun 23. Cerebrovasc Dis. 2004. PMID: 15218279 Review.
- How to write an original radiological research manuscript. Bannas P, Reeder SB. Bannas P, et al. Eur Radiol. 2017 Nov;27(11):4455-4460. doi: 10.1007/s00330-017-4879-8. Epub 2017 Jun 14. Eur Radiol. 2017. PMID: 28616726 Free PMC article. Review.
- Essential Guide to Manuscript Writing for Academic Dummies: An Editor's Perspective. Aga SS, Nissar S. Aga SS, et al. Biochem Res Int. 2022 Sep 1;2022:1492058. doi: 10.1155/2022/1492058. eCollection 2022. Biochem Res Int. 2022. PMID: 36092536 Free PMC article. Review.
- Search in MeSH
LinkOut - more resources
Full text sources.
- Ovid Technologies, Inc.
- Wolters Kluwer

- Citation Manager
NCBI Literature Resources
MeSH PMC Bookshelf Disclaimer
The PubMed wordmark and PubMed logo are registered trademarks of the U.S. Department of Health and Human Services (HHS). Unauthorized use of these marks is strictly prohibited.
- How it works

How to Write the Dissertation Findings or Results – Tips
Published by Grace Graffin at August 11th, 2021 , Revised On August 13, 2024
Each part of the dissertation is unique, and some general and specific rules must be followed. The dissertation’s findings section presents the key results of your research without interpreting their meaning .
Theoretically, this is an exciting section of a dissertation because it involves writing what you have observed and found. However, it can be a little tricky if there is too much information to confuse the readers.
The goal is to include only the essential and relevant findings in this section. The results must be presented in an orderly sequence to provide clarity to the readers.
This section of the dissertation should be easy for the readers to follow, so you should avoid going into a lengthy debate over the interpretation of the results.
It is vitally important to focus only on clear and precise observations. The findings chapter of the dissertation is theoretically the easiest to write.
It includes statistical analysis and a brief write-up about whether or not the results emerging from the analysis are significant. This segment should be written in the past sentence as you describe what you have done in the past.
This article will provide detailed information about how to write the findings of a dissertation .
When to Write Dissertation Findings Chapter
As soon as you have gathered and analysed your data, you can start to write up the findings chapter of your dissertation paper. Remember that it is your chance to report the most notable findings of your research work and relate them to the research hypothesis or research questions set out in the introduction chapter of the dissertation .
You will be required to separately report your study’s findings before moving on to the discussion chapter if your dissertation is based on the collection of primary data or experimental work.
However, you may not be required to have an independent findings chapter if your dissertation is purely descriptive and focuses on the analysis of case studies or interpretation of texts.
- Always report the findings of your research in the past tense.
- The dissertation findings chapter varies from one project to another, depending on the data collected and analyzed.
- Avoid reporting results that are not relevant to your research questions or research hypothesis.
Does your Dissertation Have the Following?
- Great Research/Sources
- Perfect Language
- Accurate Sources
If not, we can help. Our panel of experts makes sure to keep the 3 pillars of the Dissertation strong.

1. Reporting Quantitative Findings
The best way to present your quantitative findings is to structure them around the research hypothesis or questions you intend to address as part of your dissertation project.
Report the relevant findings for each research question or hypothesis, focusing on how you analyzed them.
Analysis of your findings will help you determine how they relate to the different research questions and whether they support the hypothesis you formulated.
While you must highlight meaningful relationships, variances, and tendencies, it is important not to guess their interpretations and implications because this is something to save for the discussion and conclusion chapters.
Any findings not directly relevant to your research questions or explanations concerning the data collection process should be added to the dissertation paper’s appendix section.
Use of Figures and Tables in Dissertation Findings
Suppose your dissertation is based on quantitative research. In that case, it is important to include charts, graphs, tables, and other visual elements to help your readers understand the emerging trends and relationships in your findings.
Repeating information will give the impression that you are short on ideas. Refer to all charts, illustrations, and tables in your writing but avoid recurrence.
The text should be used only to elaborate and summarize certain parts of your results. On the other hand, illustrations and tables are used to present multifaceted data.
It is recommended to give descriptive labels and captions to all illustrations used so the readers can figure out what each refers to.
How to Report Quantitative Findings
Here is an example of how to report quantitative results in your dissertation findings chapter;
Two hundred seventeen participants completed both the pretest and post-test and a Pairwise T-test was used for the analysis. The quantitative data analysis reveals a statistically significant difference between the mean scores of the pretest and posttest scales from the Teachers Discovering Computers course. The pretest mean was 29.00 with a standard deviation of 7.65, while the posttest mean was 26.50 with a standard deviation of 9.74 (Table 1). These results yield a significance level of .000, indicating a strong treatment effect (see Table 3). With the correlation between the scores being .448, the little relationship is seen between the pretest and posttest scores (Table 2). This leads the researcher to conclude that the impact of the course on the educators’ perception and integration of technology into the curriculum is dramatic.
Paired Samples
| Mean | N | Std. Deviation | Std. Error Mean | ||
|---|---|---|---|---|---|
| PRESCORE | 29.00 | 217 | 7.65 | .519 | |
| PSTSCORE | 26.00 | 217 | 9.74 | .661 |
Paired Samples Correlation
| N | Correlation | Sig. | ||
|---|---|---|---|---|
| PRESCORE & PSTSCORE | 217 | .448 | .000 |
Paired Samples Test
| Paired Differences | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Mean | Std. Deviation | Std. Error Mean | 95% Confidence Interval of the Difference | t | df | Sig. (2-tailed) | |||
| Lower | Upper | ||||||||
| Pair 1 | PRESCORE-PSTSCORE | 2.50 | 9.31 | .632 | 1.26 | 3.75 | 3.967 | 216 | .000 |
Also Read: How to Write the Abstract for the Dissertation.
2. Reporting Qualitative Findings
A notable issue with reporting qualitative findings is that not all results directly relate to your research questions or hypothesis.
The best way to present the results of qualitative research is to frame your findings around the most critical areas or themes you obtained after you examined the data.
In-depth data analysis will help you observe what the data shows for each theme. Any developments, relationships, patterns, and independent responses directly relevant to your research question or hypothesis should be mentioned to the readers.
Additional information not directly relevant to your research can be included in the appendix .
How to Report Qualitative Findings
Here is an example of how to report qualitative results in your dissertation findings chapter;
The last question of the interview focused on the need for improvement in Thai ready-to-eat products and the industry at large, emphasizing the need for enhancement in the current products being offered in the market. When asked if there was any particular need for Thai ready-to-eat meals to be improved and how to improve them in case of ‘yes,’ the males replied mainly by saying that the current products need improvement in terms of the use of healthier raw materials and preservatives or additives. There was an agreement amongst all males concerning the need to improve the industry for ready-to-eat meals and the use of more healthy items to prepare such meals. The females were also of the opinion that the fast-food items needed to be improved in the sense that more healthy raw materials such as vegetable oil and unsaturated fats, including whole-wheat products, to overcome risks associated with trans fat leading to obesity and hypertension should be used for the production of RTE products. The frozen RTE meals and packaged snacks included many preservatives and chemical-based flavouring enhancers that harmed human health and needed to be reduced. The industry is said to be aware of this fact and should try to produce RTE products that benefit the community in terms of healthy consumption.
Looking for dissertation help?
Research prospect to the rescue then.
We have expert writers on our team who are skilled at helping students with dissertations across a variety of disciplines. Guaranteeing 100% satisfaction!

What to Avoid in Dissertation Findings Chapter
- Avoid using interpretive and subjective phrases and terms such as “confirms,” “reveals,” “suggests,” or “validates.” These terms are more suitable for the discussion chapter , where you will be expected to interpret the results in detail.
- Only briefly explain findings in relation to the key themes, hypothesis, and research questions. You don’t want to write a detailed subjective explanation for any research questions at this stage.
The Do’s of Writing the Findings or Results Section
- Ensure you are not presenting results from other research studies in your findings.
- Observe whether or not your hypothesis is tested or research questions answered.
- Illustrations and tables present data and are labelled to help your readers understand what they relate to.
- Use software such as Excel, STATA, and SPSS to analyse results and important trends.
Essential Guidelines on How to Write Dissertation Findings
The dissertation findings chapter should provide the context for understanding the results. The research problem should be repeated, and the research goals should be stated briefly.
This approach helps to gain the reader’s attention toward the research problem. The first step towards writing the findings is identifying which results will be presented in this section.
The results relevant to the questions must be presented, considering whether the results support the hypothesis. You do not need to include every result in the findings section. The next step is ensuring the data can be appropriately organized and accurate.
You will need to have a basic idea about writing the findings of a dissertation because this will provide you with the knowledge to arrange the data chronologically.
Start each paragraph by writing about the most important results and concluding the section with the most negligible actual results.
A short paragraph can conclude the findings section, summarising the findings so readers will remember as they transition to the next chapter. This is essential if findings are unexpected or unfamiliar or impact the study.
Our writers can help you with all parts of your dissertation, including statistical analysis of your results . To obtain free non-binding quotes, please complete our online quote form here .
Be Impartial in your Writing
When crafting your findings, knowing how you will organize the work is important. The findings are the story that needs to be told in response to the research questions that have been answered.
Therefore, the story needs to be organized to make sense to you and the reader. The findings must be compelling and responsive to be linked to the research questions being answered.
Always ensure that the size and direction of any changes, including percentage change, can be mentioned in the section. The details of p values or confidence intervals and limits should be included.
The findings sections only have the relevant parts of the primary evidence mentioned. Still, it is a good practice to include all the primary evidence in an appendix that can be referred to later.
The results should always be written neutrally without speculation or implication. The statement of the results mustn’t have any form of evaluation or interpretation.
Negative results should be added in the findings section because they validate the results and provide high neutrality levels.
The length of the dissertation findings chapter is an important question that must be addressed. It should be noted that the length of the section is directly related to the total word count of your dissertation paper.
The writer should use their discretion in deciding the length of the findings section or refer to the dissertation handbook or structure guidelines.
It should neither belong nor be short nor concise and comprehensive to highlight the reader’s main findings.
Ethically, you should be confident in the findings and provide counter-evidence. Anything that does not have sufficient evidence should be discarded. The findings should respond to the problem presented and provide a solution to those questions.
Structure of the Findings Chapter
The chapter should use appropriate words and phrases to present the results to the readers. Logical sentences should be used, while paragraphs should be linked to produce cohesive work.
You must ensure all the significant results have been added in the section. Recheck after completing the section to ensure no mistakes have been made.
The structure of the findings section is something you may have to be sure of primarily because it will provide the basis for your research work and ensure that the discussions section can be written clearly and proficiently.
One way to arrange the results is to provide a brief synopsis and then explain the essential findings. However, there should be no speculation or explanation of the results, as this will be done in the discussion section.
Another way to arrange the section is to present and explain a result. This can be done for all the results while the section is concluded with an overall synopsis.
This is the preferred method when you are writing more extended dissertations. It can be helpful when multiple results are equally significant. A brief conclusion should be written to link all the results and transition to the discussion section.
Numerous data analysis dissertation examples are available on the Internet, which will help you improve your understanding of writing the dissertation’s findings.
Problems to Avoid When Writing Dissertation Findings
One of the problems to avoid while writing the dissertation findings is reporting background information or explaining the findings. This should be done in the introduction section .
You can always revise the introduction chapter based on the data you have collected if that seems an appropriate thing to do.
Raw data or intermediate calculations should not be added in the findings section. Always ask your professor if raw data needs to be included.
If the data is to be included, then use an appendix or a set of appendices referred to in the text of the findings chapter.
Do not use vague or non-specific phrases in the findings section. It is important to be factual and concise for the reader’s benefit.
The findings section presents the crucial data collected during the research process. It should be presented concisely and clearly to the reader. There should be no interpretation, speculation, or analysis of the data.
The significant results should be categorized systematically with the text used with charts, figures, and tables. Furthermore, avoiding using vague and non-specific words in this section is essential.
It is essential to label the tables and visual material properly. You should also check and proofread the section to avoid mistakes.
The dissertation findings chapter is a critical part of your overall dissertation paper. If you struggle with presenting your results and statistical analysis, our expert dissertation writers can help you get things right. Whether you need help with the entire dissertation paper or individual chapters, our dissertation experts can provide customized dissertation support .
FAQs About Findings of a Dissertation
How do i report quantitative findings.
The best way to present your quantitative findings is to structure them around the research hypothesis or research questions you intended to address as part of your dissertation project. Report the relevant findings for each of the research questions or hypotheses, focusing on how you analyzed them.
How do I report qualitative findings?
The best way to present the qualitative research results is to frame your findings around the most important areas or themes that you obtained after examining the data.
An in-depth analysis of the data will help you observe what the data is showing for each theme. Any developments, relationships, patterns, and independent responses that are directly relevant to your research question or hypothesis should be clearly mentioned for the readers.
Can I use interpretive phrases like ‘it confirms’ in the finding chapter?
No, It is highly advisable to avoid using interpretive and subjective phrases in the finding chapter. These terms are more suitable for the discussion chapter , where you will be expected to provide your interpretation of the results in detail.
Can I report the results from other research papers in my findings chapter?
NO, you must not be presenting results from other research studies in your findings.
You May Also Like
Finding it difficult to maintain a good relationship with your supervisor? Here are some tips on ‘How to Deal with an Unhelpful Dissertation Supervisor’.
Dissertation Methodology is the crux of dissertation project. In this article, we will provide tips for you to write an amazing dissertation methodology.
Learn how to write a good declaration page for your thesis with the help of our step-by-step comprehensive guide. Read now.
USEFUL LINKS
LEARNING RESOURCES

COMPANY DETAILS

- How It Works

How To Write The Results/Findings Chapter
For qualitative studies (dissertations & theses).
By: Jenna Crossley (PhD). Expert Reviewed By: Dr. Eunice Rautenbach | August 2021
So, you’ve collected and analysed your qualitative data, and it’s time to write up your results chapter. But where do you start? In this post, we’ll guide you through the qualitative results chapter (also called the findings chapter), step by step.
Overview: Qualitative Results Chapter
- What (exactly) the qualitative results chapter is
- What to include in your results chapter
- How to write up your results chapter
- A few tips and tricks to help you along the way
- Free results chapter template
What exactly is the results chapter?
The results chapter in a dissertation or thesis (or any formal academic research piece) is where you objectively and neutrally present the findings of your qualitative analysis (or analyses if you used multiple qualitative analysis methods ). This chapter can sometimes be combined with the discussion chapter (where you interpret the data and discuss its meaning), depending on your university’s preference. We’ll treat the two chapters as separate, as that’s the most common approach.
In contrast to a quantitative results chapter that presents numbers and statistics, a qualitative results chapter presents data primarily in the form of words . But this doesn’t mean that a qualitative study can’t have quantitative elements – you could, for example, present the number of times a theme or topic pops up in your data, depending on the analysis method(s) you adopt.
Adding a quantitative element to your study can add some rigour, which strengthens your results by providing more evidence for your claims. This is particularly common when using qualitative content analysis. Keep in mind though that qualitative research aims to achieve depth, richness and identify nuances , so don’t get tunnel vision by focusing on the numbers. They’re just cream on top in a qualitative analysis.
So, to recap, the results chapter is where you objectively present the findings of your analysis, without interpreting them (you’ll save that for the discussion chapter). With that out the way, let’s take a look at what you should include in your results chapter.

What should you include in the results chapter?
As we’ve mentioned, your qualitative results chapter should purely present and describe your results , not interpret them in relation to the existing literature or your research questions . Any speculations or discussion about the implications of your findings should be reserved for your discussion chapter.
In your results chapter, you’ll want to talk about your analysis findings and whether or not they support your hypotheses (if you have any). Naturally, the exact contents of your results chapter will depend on which qualitative analysis method (or methods) you use. For example, if you were to use thematic analysis, you’d detail the themes identified in your analysis, using extracts from the transcripts or text to support your claims.
While you do need to present your analysis findings in some detail, you should avoid dumping large amounts of raw data in this chapter. Instead, focus on presenting the key findings and using a handful of select quotes or text extracts to support each finding . The reams of data and analysis can be relegated to your appendices.
While it’s tempting to include every last detail you found in your qualitative analysis, it is important to make sure that you report only that which is relevant to your research aims, objectives and research questions . Always keep these three components, as well as your hypotheses (if you have any) front of mind when writing the chapter and use them as a filter to decide what’s relevant and what’s not.
Need a helping hand?
How do I write the results chapter?
Now that we’ve covered the basics, it’s time to look at how to structure your chapter. Broadly speaking, the results chapter needs to contain three core components – the introduction, the body and the concluding summary. Let’s take a look at each of these.
Section 1: Introduction
The first step is to craft a brief introduction to the chapter. This intro is vital as it provides some context for your findings. In your introduction, you should begin by reiterating your problem statement and research questions and highlight the purpose of your research . Make sure that you spell this out for the reader so that the rest of your chapter is well contextualised.
The next step is to briefly outline the structure of your results chapter. In other words, explain what’s included in the chapter and what the reader can expect. In the results chapter, you want to tell a story that is coherent, flows logically, and is easy to follow , so make sure that you plan your structure out well and convey that structure (at a high level), so that your reader is well oriented.
The introduction section shouldn’t be lengthy. Two or three short paragraphs should be more than adequate. It is merely an introduction and overview, not a summary of the chapter.
Pro Tip – To help you structure your chapter, it can be useful to set up an initial draft with (sub)section headings so that you’re able to easily (re)arrange parts of your chapter. This will also help your reader to follow your results and give your chapter some coherence. Be sure to use level-based heading styles (e.g. Heading 1, 2, 3 styles) to help the reader differentiate between levels visually. You can find these options in Word (example below).
Section 2: Body
Before we get started on what to include in the body of your chapter, it’s vital to remember that a results section should be completely objective and descriptive, not interpretive . So, be careful not to use words such as, “suggests” or “implies”, as these usually accompany some form of interpretation – that’s reserved for your discussion chapter.
The structure of your body section is very important , so make sure that you plan it out well. When planning out your qualitative results chapter, create sections and subsections so that you can maintain the flow of the story you’re trying to tell. Be sure to systematically and consistently describe each portion of results. Try to adopt a standardised structure for each portion so that you achieve a high level of consistency throughout the chapter.
For qualitative studies, results chapters tend to be structured according to themes , which makes it easier for readers to follow. However, keep in mind that not all results chapters have to be structured in this manner. For example, if you’re conducting a longitudinal study, you may want to structure your chapter chronologically. Similarly, you might structure this chapter based on your theoretical framework . The exact structure of your chapter will depend on the nature of your study , especially your research questions.
As you work through the body of your chapter, make sure that you use quotes to substantiate every one of your claims . You can present these quotes in italics to differentiate them from your own words. A general rule of thumb is to use at least two pieces of evidence per claim, and these should be linked directly to your data. Also, remember that you need to include all relevant results , not just the ones that support your assumptions or initial leanings.
In addition to including quotes, you can also link your claims to the data by using appendices , which you should reference throughout your text. When you reference, make sure that you include both the name/number of the appendix , as well as the line(s) from which you drew your data.
As referencing styles can vary greatly, be sure to look up the appendix referencing conventions of your university’s prescribed style (e.g. APA , Harvard, etc) and keep this consistent throughout your chapter.
Section 3: Concluding summary
The concluding summary is very important because it summarises your key findings and lays the foundation for the discussion chapter . Keep in mind that some readers may skip directly to this section (from the introduction section), so make sure that it can be read and understood well in isolation.
In this section, you need to remind the reader of the key findings. That is, the results that directly relate to your research questions and that you will build upon in your discussion chapter. Remember, your reader has digested a lot of information in this chapter, so you need to use this section to remind them of the most important takeaways.
Importantly, the concluding summary should not present any new information and should only describe what you’ve already presented in your chapter. Keep it concise – you’re not summarising the whole chapter, just the essentials.
Tips for writing an A-grade results chapter
Now that you’ve got a clear picture of what the qualitative results chapter is all about, here are some quick tips and reminders to help you craft a high-quality chapter:
- Your results chapter should be written in the past tense . You’ve done the work already, so you want to tell the reader what you found , not what you are currently finding .
- Make sure that you review your work multiple times and check that every claim is adequately backed up by evidence . Aim for at least two examples per claim, and make use of an appendix to reference these.
- When writing up your results, make sure that you stick to only what is relevant . Don’t waste time on data that are not relevant to your research objectives and research questions.
- Use headings and subheadings to create an intuitive, easy to follow piece of writing. Make use of Microsoft Word’s “heading styles” and be sure to use them consistently.
- When referring to numerical data, tables and figures can provide a useful visual aid. When using these, make sure that they can be read and understood independent of your body text (i.e. that they can stand-alone). To this end, use clear, concise labels for each of your tables or figures and make use of colours to code indicate differences or hierarchy.
- Similarly, when you’re writing up your chapter, it can be useful to highlight topics and themes in different colours . This can help you to differentiate between your data if you get a bit overwhelmed and will also help you to ensure that your results flow logically and coherently.
If you have any questions, leave a comment below and we’ll do our best to help. If you’d like 1-on-1 help with your results chapter (or any chapter of your dissertation or thesis), check out our private dissertation coaching service here or book a free initial consultation to discuss how we can help you.

Psst... there’s more!
This post was based on one of our popular Research Bootcamps . If you're working on a research project, you'll definitely want to check this out ...
22 Comments
This was extremely helpful. Thanks a lot guys
Hi, thanks for the great research support platform created by the gradcoach team!
I wanted to ask- While “suggests” or “implies” are interpretive terms, what terms could we use for the results chapter? Could you share some examples of descriptive terms?
I think that instead of saying, ‘The data suggested, or The data implied,’ you can say, ‘The Data showed or revealed, or illustrated or outlined’…If interview data, you may say Jane Doe illuminated or elaborated, or Jane Doe described… or Jane Doe expressed or stated.
I found this article very useful. Thank you very much for the outstanding work you are doing.
What if i have 3 different interviewees answering the same interview questions? Should i then present the results in form of the table with the division on the 3 perspectives or rather give a results in form of the text and highlight who said what?
I think this tabular representation of results is a great idea. I am doing it too along with the text. Thanks
That was helpful was struggling to separate the discussion from the findings
this was very useful, Thank you.
Very helpful, I am confident to write my results chapter now.
It is so helpful! It is a good job. Thank you very much!
Very useful, well explained. Many thanks.
Hello, I appreciate the way you provided a supportive comments about qualitative results presenting tips
I loved this! It explains everything needed, and it has helped me better organize my thoughts. What words should I not use while writing my results section, other than subjective ones.
Thanks a lot, it is really helpful
Thank you so much dear, i really appropriate your nice explanations about this.
Thank you so much for this! I was wondering if anyone could help with how to prproperly integrate quotations (Excerpts) from interviews in the finding chapter in a qualitative research. Please GradCoach, address this issue and provide examples.
what if I’m not doing any interviews myself and all the information is coming from case studies that have already done the research.
Very helpful thank you.
This was very helpful as I was wondering how to structure this part of my dissertation, to include the quotes… Thanks for this explanation
This is very helpful, thanks! I am required to write up my results chapters with the discussion in each of them – any tips and tricks for this strategy?
For qualitative studies, can the findings be structured according to the Research questions? Thank you.
Do I need to include literature/references in my findings chapter?
Submit a Comment Cancel reply
Your email address will not be published. Required fields are marked *
Save my name, email, and website in this browser for the next time I comment.
- Print Friendly

- Chapter Seven: Presenting Your Results
This chapter serves as the culmination of the previous chapters, in that it focuses on how to present the results of one's study, regardless of the choice made among the three methods. Writing in academics has a form and style that you will want to apply not only to report your own research, but also to enhance your skills at reading original research published in academic journals. Beyond the basic academic style of report writing, there are specific, often unwritten assumptions about how quantitative, qualitative, and critical/rhetorical studies should be organized and the information they should contain. This chapter discusses how to present your results in writing, how to write accessibly, how to visualize data, and how to present your results in person.
- Chapter One: Introduction
- Chapter Two: Understanding the distinctions among research methods
- Chapter Three: Ethical research, writing, and creative work
- Chapter Four: Quantitative Methods (Part 1)
- Chapter Four: Quantitative Methods (Part 2 - Doing Your Study)
- Chapter Four: Quantitative Methods (Part 3 - Making Sense of Your Study)
- Chapter Five: Qualitative Methods (Part 1)
- Chapter Five: Qualitative Data (Part 2)
- Chapter Six: Critical / Rhetorical Methods (Part 1)
- Chapter Six: Critical / Rhetorical Methods (Part 2)
Written Presentation of Results
Once you've gone through the process of doing communication research – using a quantitative, qualitative, or critical/rhetorical methodological approach – the final step is to communicate it.
The major style manuals (the APA Manual, the MLA Handbook, and Turabian) are very helpful in documenting the structure of writing a study, and are highly recommended for consultation. But, no matter what style manual you may use, there are some common elements to the structure of an academic communication research paper.
Title Page :
This is simple: Your Paper's Title, Your Name, Your Institutional Affiliation (e.g., University), and the Date, each on separate lines, centered on the page. Try to make your title both descriptive (i.e., it gives the reader an idea what the study is about) and interesting (i.e., it is catchy enough to get one's attention).
For example, the title, "The uncritical idealization of a compensated psychopath character in a popular book series," would not be an inaccurate title for a published study, but it is rather vague and exceedingly boring. That study's author fortunately chose the title, "A boyfriend to die for: Edward Cullen as compensated psychopath in Stephanie Meyer's Twilight ," which is more precisely descriptive, and much more interesting (Merskin, 2011). The use of the colon in academic titles can help authors accomplish both objectives: a catchy but relevant phrase, followed by a more clear explanation of the article's topic.
In some instances, you might be asked to write an abstract, which is a summary of your paper that can range in length from 75 to 250 words. If it is a published paper, it is useful to include key search terms in this brief description of the paper (the title may already have a few of these terms as well). Although this may be the last thing your write, make it one of the best things you write, because this may be the first thing your audience reads about the paper (and may be the only thing read if it is written badly). Summarize the problem/research question, your methodological approach, your results and conclusions, and the significance of the paper in the abstract.
Quantitative and qualitative studies will most typically use the rest of the section titles noted below. Critical/rhetorical studies will include many of the same steps, but will often have different headings. For example, a critical/rhetorical paper will have an introduction, definition of terms, and literature review, followed by an analysis (often divided into sections by areas of investigation) and ending with a conclusion/implications section. Because critical/rhetorical research is much more descriptive, the subheadings in such a paper are often times not generic subheads like "literature review," but instead descriptive subheadings that apply to the topic at hand, as seen in the schematic below. Because many journals expect the article to follow typical research paper headings of introduction, literature review, methods, results, and discussion, we discuss these sections briefly next.
Introduction:
As you read social scientific journals (see chapter 1 for examples), you will find that they tend to get into the research question quickly and succinctly. Journal articles from the humanities tradition tend to be more descriptive in the introduction. But, in either case, it is good to begin with some kind of brief anecdote that gets the reader engaged in your work and lets the reader understand why this is an interesting topic. From that point, state your research question, define the problem (see Chapter One) with an overview of what we do and don't know, and finally state what you will do, or what you want to find out. The introduction thus builds the case for your topic, and is the beginning of building your argument, as we noted in chapter 1.
By the end of the Introduction, the reader should know what your topic is, why it is a significant communication topic, and why it is necessary that you investigate it (e.g., it could be there is gap in literature, you will conduct valuable exploratory research, or you will provide a new model for solving some professional or social problem).
Literature Review:
The literature review summarizes and organizes the relevant books, articles, and other research in this area. It sets up both quantitative and qualitative studies, showing the need for the study. For critical/rhetorical research, the literature review often incorporates the description of the historical context and heuristic vocabulary, with key terms defined in this section of the paper. For more detail on writing a literature review, see Appendix 1.
The methods of your paper are the processes that govern your research, where the researcher explains what s/he did to solve the problem. As you have seen throughout this book, in communication studies, there are a number of different types of research methods. For example, in quantitative research, one might conduct surveys, experiments, or content analysis. In qualitative research, one might instead use interviews and observations. Critical/rhetorical studies methods are more about the interpretation of texts or the study of popular culture as communication. In creative communication research, the method may be an interpretive performance studies or filmmaking. Other methods used sometimes alone, or in combination with other methods, include legal research, historical research, and political economy research.
In quantitative and qualitative research papers, the methods will be most likely described according to the APA manual standards. At the very least, the methods will include a description of participants, data collection, and data analysis, with specific details on each of these elements. For example, in an experiment, the researcher will describe the number of participants, the materials used, the design of the experiment, the procedure of the experiment, and what statistics will be used to address the hypotheses/research questions.
Critical/rhetorical researchers rarely have a specific section called "methods," as opposed to quantitative and qualitative researchers, but rather demonstrate the method they use for analysis throughout the writing of their piece.
Helping your reader understand the methods you used for your study is important not only for your own study's credibility, but also for possible replication of your study by other researchers. A good guideline to keep in mind is transparency . You want to be as clear as possible in describing the decisions you made in designing your study, gathering and analyzing your data so that the reader can retrace your steps and understand how you came to the conclusions you formed. A research study can be very good, but if it is not clearly described so that others can see how the results were determined or obtained, then the quality of the study and its potential contributions are lost.
After you completed your study, your findings will be listed in the results section. Particularly in a quantitative study, the results section is for revisiting your hypotheses and reporting whether or not your results supported them, and the statistical significance of the results. Whether your study supported or contradicted your hypotheses, it's always helpful to fully report what your results were. The researcher usually organizes the results of his/her results section by research question or hypothesis, stating the results for each one, using statistics to show how the research question or hypothesis was answered in the study.
The qualitative results section also may be organized by research question, but usually is organized by themes which emerged from the data collected. The researcher provides rich details from her/his observations and interviews, with detailed quotations provided to illustrate the themes identified. Sometimes the results section is combined with the discussion section.
Critical/rhetorical researchers would include their analysis often with different subheadings in what would be considered a "results" section, yet not labeled specifically this way.
Discussion:
In the discussion section, the researcher gives an appraisal of the results. Here is where the researcher considers the results, particularly in light of the literature review, and explains what the findings mean. If the results confirmed or corresponded with the findings of other literature, then that should be stated. If the results didn't support the findings of previous studies, then the researcher should develop an explanation of why the study turned out this way. Sometimes, this section is called a "conclusion" by researchers.
References:
In this section, all of the literature cited in the text should have full references in alphabetical order. Appendices: Appendix material includes items like questionnaires used in the study, photographs, documents, etc. An alphabetical letter is assigned for each piece (e.g. Appendix A, Appendix B), with a second line of title describing what the appendix contains (e.g. Participant Informed Consent, or New York Times Speech Coverage). They should be organized consistently with the order in which they are referenced in the text of the paper. The page numbers for appendices are consecutive with the paper and reference list.
Tables/Figures:
Tables and figures are referenced in the text, but included at the end of the study and numbered consecutively. (Check with your professor; some like to have tables and figures inserted within the paper's main text.) Tables generally are data in a table format, whereas figures are diagrams (such as a pie chart) and drawings (such as a flow chart).
Accessible Writing
As you may have noticed, academic writing does have a language (e.g., words like heuristic vocabulary and hypotheses) and style (e.g., literature reviews) all its own. It is important to engage in that language and style, and understand how to use it to communicate effectively in an academic context . Yet, it is also important to remember that your analyses and findings should also be written to be accessible. Writers should avoid excessive jargon, or—even worse—deploying jargon to mask an incomplete understanding of a topic.
The scourge of excessive jargon in academic writing was the target of a famous hoax in 1996. A New York University physics professor submitted an article, " Transgressing the Boundaries: Toward a Transformative Hermeneutics of Quantum Gravity ," to a special issue of the academic journal Social Text devoted to science and postmodernism. The article was designed to point out how dense academic jargon can sometimes mask sloppy thinking. As the professor, Alan Sokal, had expected, the article was published. One sample sentence from the article reads:
It has thus become increasingly apparent that physical "reality", no less than social "reality", is at bottom a social and linguistic construct; that scientific "knowledge", far from being objective, reflects and encodes the dominant ideologies and power relations of the culture that produced it; that the truth claims of science are inherently theory-laden and self-referential; and consequently, that the discourse of the scientific community, for all its undeniable value, cannot assert a privileged epistemological status with respect to counter-hegemonic narratives emanating from dissident or marginalized communities. (Sokal, 1996. pp. 217-218)
According to the journal's editor, about six reviewers had read the article but didn't suspect that it was phony. A public debate ensued after Sokal revealed his hoax. Sokal said he worried that jargon and intellectual fads cause academics to lose contact with the real world and "undermine the prospect for progressive social critique" ( Scott, 1996 ). The APA Manual recommends to avoid using technical vocabulary where it is not needed or relevant or if the technical language is overused, thus becoming jargon. In short, the APA argues that "scientific jargon...grates on the reader, encumbers the communication of information, and wastes space" (American Psychological Association, 2010, p. 68).
Data Visualization
Images and words have long existed on the printed page of manuscripts, yet, until recently, relatively few researchers possessed the resources to effectively combine images combined with words (Tufte, 1990, 1983). Communication scholars are only now becoming aware of this dimension in research as computer technologies have made it possible for many people to produce and publish multimedia presentations.
Although visuals may seem to be anathema to the primacy of the written word in research, they are a legitimate way, and at times the best way, to present ideas. Visual scholar Lester Faigley et al. (2004) explains how data visualizations have become part of our daily lives:
Visualizations can shed light on research as well. London-based David McCandless specializes in visualizing interesting research questions, or in his words "the questions I wanted answering" (2009, p. 7). His images include a graph of the peak times of the year for breakups (based on Facebook status updates), a radiation dosage chart , and some experiments with the Google Ngram Viewer , which charts the appearance of keywords in millions of books over hundreds of years.
The public domain image below creatively maps U.S. Census data of the outflow of people from California to other states between 1995 and 2000.
Visualizing one's research is possible in multiple ways. A simple technology, for example, is to enter data into a spreadsheet such as Excel, and select Charts or SmartArt to generate graphics. A number of free web tools can also transform raw data into useful charts and graphs. Many Eyes , an open source data visualization tool (sponsored by IBM Research), says its goal "is to 'democratize' visualization and to enable a new social kind of data analysis" (IBM, 2011). Another tool, Soundslides , enables users to import images and audio to create a photographic slideshow, while the program handles all of the background code. Other tools, often open source and free, can help visual academic research into interactive maps; interactive, image-based timelines; interactive charts; and simple 2-D and 3-D animations. Adobe Creative Suite (which includes popular software like Photoshop) is available on most computers at universities, but open source alternatives exist as well. Gimp is comparable to Photoshop, and it is free and relatively easy to use.
One online performance studies journal, Liminalities , is an excellent example of how "research" can be more than just printed words. In each issue, traditional academic essays and book reviews are often supported photographs, while other parts of an issue can include video, audio, and multimedia contributions. The journal, founded in 2005, treats performance itself as a methodology, and accepts contribution in html, mp3, Quicktime, and Flash formats.
For communication researchers, there is also a vast array of visual digital archives available online. Many of these archives are located at colleges and universities around the world, where digital librarians are spearheading a massive effort to make information—print, audio, visual, and graphic—available to the public as part of a global information commons. For example, the University of Iowa has a considerable digital archive including historical photos documenting American railroads and a database of images related to geoscience. The University of Northern Iowa has a growing Special Collections Unit that includes digital images of every UNI Yearbook between 1905 and 1923 and audio files of UNI jazz band performances. Researchers at he University of Michigan developed OAIster , a rich database that has joined thousands of digital archives in one searchable interface. Indeed, virtually every academic library is now digitizing all types of media, not just texts, and making them available for public viewing and, when possible, for use in presenting research. In addition to academic collections, the Library of Congress and the National Archives offer an ever-expanding range of downloadable media; commercial, user-generated databases such as Flickr, Buzznet, YouTube and Google Video offer a rich resource of images that are often free of copyright constraints (see Chapter 3 about Creative Commons licenses) and nonprofit endeavors, such as the Internet Archive , contain a formidable collection of moving images, still photographs, audio files (including concert recordings), and open source software.
Presenting your Work in Person
As Communication students, it's expected that you are not only able to communicate your research project in written form but also in person.
Before you do any oral presentation, it's good to have a brief "pitch" ready for anyone who asks you about your research. The pitch is routine in Hollywood: a screenwriter has just a few minutes to present an idea to a producer. Although your pitch will be more sophisticated than, say, " Snakes on a Plane " (which unfortunately was made into a movie), you should in just a few lines be able to explain the gist of your research to anyone who asks. Developing this concise description, you will have some practice in distilling what might be a complicated topic into one others can quickly grasp.
Oral presentation
In most oral presentations of research, whether at the end of a semester, or at a research symposium or conference, you will likely have just 10 to 20 minutes. This is probably not enough time to read the entire paper aloud, which is not what you should do anyway if you want people to really listen (although, unfortunately some make this mistake). Instead, the point of the presentation should be to present your research in an interesting manner so the listeners will want to read the whole thing. In the presentation, spend the least amount of time on the literature review (a very brief summary will suffice) and the most on your own original contribution. In fact, you may tell your audience that you are only presenting on one portion of the paper, and that you would be happy to talk more about your research and findings in the question and answer session that typically follows. Consider your presentation the beginning of a dialogue between you and the audience. Your tone shouldn't be "I have found everything important there is to find, and I will cram as much as I can into this presentation," but instead "I found some things you will find interesting, but I realize there is more to find."
Turabian (2007) has a helpful chapter on presenting research. Most important, she emphasizes, is to remember that your audience members are listeners, not readers. Thus, recall the lessons on speech making in your college oral communication class. Give an introduction, tell them what the problem is, and map out what you will present to them. Organize your findings into a few points, and don't get bogged down in minutiae. (The minutiae are for readers to find if they wish, not for listeners to struggle through.) PowerPoint slides are acceptable, but don't read them. Instead, create an outline of a few main points, and practice your presentation.
Turabian suggests an introduction of not more than three minutes, which should include these elements:
- The research topic you will address (not more than a minute).
- Your research question (30 seconds or less)
- An answer to "so what?" – explaining the relevance of your research (30 seconds)
- Your claim, or argument (30 seconds or less)
- The map of your presentation structure (30 seconds or less)
As Turabian (2007) suggests, "Rehearse your introduction, not only to get it right, but to be able to look your audience in the eye as you give it. You can look down at notes later" (p. 125).
Poster presentation
In some symposiums and conferences, you may be asked to present at a "poster" session. Instead of presenting on a panel of 4-5 people to an audience, a poster presenter is with others in a large hall or room, and talks one-on-one with visitors who look at the visual poster display of the research. As in an oral presentation, a poster highlights just the main point of the paper. Then, if visitors have questions, the author can informally discuss her/his findings.
To attract attention, poster presentations need to be nicely designed, or in the words of an advertising professor who schedules poster sessions at conferences, "be big, bold, and brief" ( Broyles , 2011). Large type (at least 18 pt.), graphics, tables, and photos are recommended.
A poster presentation session at a conference, by David Eppstein (Own work) [CC-BY-SA-3.0 ( www.creativecommons.org/licenses/by-sa/3.0 )], via Wikimedia Commons]
The Association for Education in Journalism and Mass Communication (AEJMC) has a template for making an effective poster presentation . Many universities, copy shops, and Internet services also have large-scale printers, to print full-color research poster designs that can be rolled up and transported in a tube.
Judging Others' Research
After taking this course, you should have a basic knowledge of research methods. There will still be some things that may mystify you as a reader of other's research. For example, you may not be able to interpret the coefficients for statistical significance, or make sense of a complex structural equation. Some specialized vocabulary may still be difficult.
But, you should understand how to critically review research. For example, imagine you have been asked to do a blind (i.e., the author's identity is concealed) "peer review" of communication research for acceptance to a conference, or publication in an academic journal. For most conferences and journals , submissions are made online, where editors can manage the flow and assign reviews to papers. The evaluations reviewers make are based on the same things that we have covered in this book. For example, the conference for the AEJMC ask reviewers to consider (on a five-point scale, from Excellent to Poor) a number of familiar research dimensions, including the paper's clarity of purpose, literature review, clarity of research method, appropriateness of research method, evidence presented clearly, evidence supportive of conclusions, general writing and organization, and the significance of the contribution to the field.
Beyond academia, it is likely you will more frequently apply the lessons of research methods as a critical consumer of news, politics, and everyday life. Just because some expert cites a number or presents a conclusion doesn't mean it's automatically true. John Allen Paulos, in his book A Mathematician reads the newspaper , suggests some basic questions we can ask. "If statistics were presented, how were they obtained? How confident can we be of them? Were they derived from a random sample or from a collection of anecdotes? Does the correlation suggest a causal relationship, or is it merely a coincidence?" (1997, p. 201).
Through the study of research methods, we have begun to build a critical vocabulary and understanding to ask good questions when others present "knowledge." For example, if Candidate X won a straw poll in Iowa, does that mean she'll get her party's nomination? If Candidate Y wins an open primary in New Hampshire, does that mean he'll be the next president? If Candidate Z sheds a tear, does it matter what the context is, or whether that candidate is a man or a woman? What we learn in research methods about validity, reliability, sampling, variables, research participants, epistemology, grounded theory, and rhetoric, we can consider whether the "knowledge" that is presented in the news is a verifiable fact, a sound argument, or just conjecture.
American Psychological Association (2010). Publication manual of the American Psychological Association (6th ed.). Washington, DC: Author.
Broyles, S. (2011). "About poster sessions." AEJMC. http://www.aejmc.org/home/2013/01/about-poster-sessions/ .
Faigley, L., George, D., Palchik, A., Selfe, C. (2004). Picturing texts . New York: W.W. Norton & Company.
IBM (2011). Overview of Many Eyes. http://www.research.ibm.com/social/projects_manyeyes.shtml .
McCandless, D. (2009). The visual miscellaneum . New York: Collins Design.
Merskin, D. (2011). A boyfriend to die for: Edward Cullen as compensated psychopath in Stephanie Meyer's Twilight. Journal of Communication Inquiry 35: 157-178. doi:10.1177/0196859911402992
Paulos, J. A. (1997). A mathematician reads the newspaper . New York: Anchor.
Scott, J. (1996, May 18). Postmodern gravity deconstructed, slyly. New York Times , http://www.nytimes.com/books/98/11/15/specials/sokal-text.html .
Sokal, A. (1996). Transgressing the boundaries: towards a transformative hermeneutics of quantum gravity. Social Text 46/47, 217-252.
Tufte, E. R. (1990). Envisioning information . Cheshire, CT: Graphics Press.
Tufte, E. R. (1983). The visual display of quantitative information . Cheshire, CT: Graphics Press.
Turabian, Kate L. (2007). A manual for writers of research papers, theses, and dissertations: Chicago style guide for students and researchers (7th ed.). Chicago: University of Chicago Press.

Organizing Academic Research Papers: 7. The Results
- Purpose of Guide
- Design Flaws to Avoid
- Glossary of Research Terms
- Narrowing a Topic Idea
- Broadening a Topic Idea
- Extending the Timeliness of a Topic Idea
- Academic Writing Style
- Choosing a Title
- Making an Outline
- Paragraph Development
- Executive Summary
- Background Information
- The Research Problem/Question
- Theoretical Framework
- Citation Tracking
- Content Alert Services
- Evaluating Sources
- Primary Sources
- Secondary Sources
- Tertiary Sources
- What Is Scholarly vs. Popular?
- Qualitative Methods
- Quantitative Methods
- Using Non-Textual Elements
- Limitations of the Study
- Common Grammar Mistakes
- Avoiding Plagiarism
- Footnotes or Endnotes?
- Further Readings
- Annotated Bibliography
- Dealing with Nervousness
- Using Visual Aids
- Grading Someone Else's Paper
- How to Manage Group Projects
- Multiple Book Review Essay
- Reviewing Collected Essays
- About Informed Consent
- Writing Field Notes
- Writing a Policy Memo
- Writing a Research Proposal
- Acknowledgements
The results section of the research paper is where you report the findings of your study based upon the information gathered as a result of the methodology [or methodologies] you applied. The results section should simply state the findings, without bias or interpretation, and arranged in a logical sequence. The results section should always be written in the past tense. A section describing results [a.k.a., "findings"] is particularly necessary if your paper includes data generated from your own research.
Importance of a Good Results Section
When formulating the results section, it's important to remember that the results of a study do not prove anything . Research results can only confirm or reject the research problem underpinning your study. However, the act of articulating the results helps you to understand the problem from within, to break it into pieces, and to view the research problem from various perspectives.
The page length of this section is set by the amount and types of data to be reported . Be concise, using non-textual elements, such as figures and tables, if appropriate, to present results more effectively. In deciding what data to describe in your results section, you must clearly distinguish material that would normally be included in a research paper from any raw data or other material that could be included as an appendix. In general, raw data should not be included in the main text of your paper unless requested to do so by your professor.
Avoid providing data that is not critical to answering the research question . The background information you described in the introduction section should provide the reader with any additional context or explanation needed to understand the results. A good rule is to always re-read the background section of your paper after you have written up your results to ensure that the reader has enough context to understand the results [and, later, how you interpreted the results in the discussion section of your paper].
Bates College; Burton, Neil et al. Doing Your Education Research Project . Los Angeles, CA: SAGE, 2008; Results . The Structure, Format, Content, and Style of a Journal-Style Scientific Paper. Department of Biology. Bates College.
Structure and Writing Style
I. Structure and Approach
For most research paper formats, there are two ways of presenting and organizing the results .
- Present the results followed by a short explanation of the findings . For example, you may have noticed an unusual correlation between two variables during the analysis of your findings. It is correct to point this out in the results section. However, speculating as to why this correlation exists, and offering a hypothesis about what may be happening, belongs in the discussion section of your paper.
- Present a section and then discuss it, before presenting the next section then discussing it, and so on . This is more common in longer papers because it helps the reader to better understand each finding. In this model, it can be helpful to provide a brief conclusion in the results section that ties each of the findings together and links to the discussion.
NOTE: The discussion section should generally follow the same format chosen in presenting and organizing the results.
II. Content
In general, the content of your results section should include the following elements:
- An introductory context for understanding the results by restating the research problem that underpins the purpose of your study.
- A summary of your key findings arranged in a logical sequence that generally follows your methodology section.
- Inclusion of non-textual elements, such as, figures, charts, photos, maps, tables, etc. to further illustrate the findings, if appropriate.
- In the text, a systematic description of your results, highlighting for the reader observations that are most relevant to the topic under investigation [remember that not all results that emerge from the methodology that you used to gather the data may be relevant].
- Use of the past tense when refering to your results.
- The page length of your results section is guided by the amount and types of data to be reported. However, focus only on findings that are important and related to addressing the research problem.
Using Non-textual Elements
- Either place figures, tables, charts, etc. within the text of the result, or include them in the back of the report--do one or the other but never do both.
- In the text, refer to each non-textual element in numbered order [e.g., Table 1, Table 2; Chart 1, Chart 2; Map 1, Map 2].
- If you place non-textual elements at the end of the report, make sure they are clearly distinguished from any attached appendix materials, such as raw data.
- Regardless of placement, each non-textual element must be numbered consecutively and complete with caption [caption goes under the figure, table, chart, etc.]
- Each non-textual element must be titled, numbered consecutively, and complete with a heading [title with description goes above the figure, table, chart, etc.].
- In proofreading your results section, be sure that each non-textual element is sufficiently complete so that it could stand on its own, separate from the text.
III. Problems to Avoid
When writing the results section, avoid doing the following :
- Discussing or interpreting your results . Save all this for the next section of your paper, although where appropriate, you should compare or contrast specific results to those found in other studies [e.g., "Similar to Smith [1990], one of the findings of this study is the strong correlation between motivation and academic achievement...."].
- Reporting background information or attempting to explain your findings ; this should have been done in your Introduction section, but don't panic! Often the results of a study point to the need to provide additional background information or to explain the topic further, so don't think you did something wrong. Revise your introduction as needed.
- Ignoring negative results . If some of your results fail to support your hypothesis, do not ignore them. Document them, then state in your discussion section why you believe a negative result emerged from your study. Note that negative results, and how you handle them, often provides you with the opportunity to write a more engaging discussion section, therefore, don't be afraid to highlight them.
- Including raw data or intermediate calculations . Ask your professor if you need to include any raw data generated by your study, such as transcripts from interviews or data files. If raw data is to be included, place it in an appendix or set of appendices that are referred to in the text.
- Be as factual and concise as possible in reporting your findings . Do not use phrases that are vague or non-specific, such as, "appeared to be greater or lesser than..." or "demonstrates promising trends that...."
- Presenting the same data or repeating the same information more than once . If you feel the need to highlight something, you will have a chance to do that in the discussion section.
- Confusing figures with tables . Be sure to properly label any non-textual elements in your paper. If you are not sure, look up the term in a dictionary.
Burton, Neil et al. Doing Your Education Research Project . Los Angeles, CA: SAGE, 2008; Caprette, David R. Writing Research Papers . Experimental Biosciences Resources. Rice University; Hancock, Dawson R. and Bob Algozzine. Doing Case Study Research: A Practical Guide for Beginning Researchers . 2nd ed. New York: Teachers College Press, 2011; Introduction to Nursing Research: Reporting Research Findings. Nursing Research: Open Access Nursing Research and Review Articles. (January 4, 2012); Reporting Research Findings. Wilder Research, in partnership with the Minnesota Department of Human Services. (February 2009); Results . The Structure, Format, Content, and Style of a Journal-Style Scientific Paper. Department of Biology. Bates College; Schafer, Mickey S. Writing the Results . Thesis Writing in the Sciences. Course Syllabus. University of Florida.
Writing Tip
Why Don't I Just Combine the Results Section with the Discussion Section?
It's not unusual to find articles in social science journals where the author(s) have combined a description of the findings from the study with a discussion about their implications. You could do this. However, if you are inexperienced writing research papers, consider creating two sections for each element in your paper as a way to better organize your thoughts and, by extension, your paper. Think of the results section as the place where you report what your study found; think of the discussion section as the place where you interpret your data and answer the "so what?" question. As you become more skilled writing research papers, you may want to meld the results of your study with a discussion of its implications.
- << Previous: Quantitative Methods
- Next: Using Non-Textual Elements >>
- Last Updated: Jul 18, 2023 11:58 AM
- URL: https://library.sacredheart.edu/c.php?g=29803
- QuickSearch
- Library Catalog
- Databases A-Z
- Publication Finder
- Course Reserves
- Citation Linker
- Digital Commons
- Our Website
Research Support
- Ask a Librarian
- Appointments
- Interlibrary Loan (ILL)
- Research Guides
- Databases by Subject
- Citation Help
Using the Library
- Reserve a Group Study Room
- Renew Books
- Honors Study Rooms
- Off-Campus Access
- Library Policies
- Library Technology
User Information
- Grad Students
- Online Students
- COVID-19 Updates
- Staff Directory
- News & Announcements
- Library Newsletter
My Accounts
- Interlibrary Loan
- Staff Site Login
FIND US ON

How to Write the Discussion Section of a Research Paper
The discussion section of a research paper analyzes and interprets the findings, provides context, compares them with previous studies, identifies limitations, and suggests future research directions.
Updated on September 15, 2023

Structure your discussion section right, and you’ll be cited more often while doing a greater service to the scientific community. So, what actually goes into the discussion section? And how do you write it?
The discussion section of your research paper is where you let the reader know how your study is positioned in the literature, what to take away from your paper, and how your work helps them. It can also include your conclusions and suggestions for future studies.
First, we’ll define all the parts of your discussion paper, and then look into how to write a strong, effective discussion section for your paper or manuscript.
Discussion section: what is it, what it does
The discussion section comes later in your paper, following the introduction, methods, and results. The discussion sets up your study’s conclusions. Its main goals are to present, interpret, and provide a context for your results.
What is it?
The discussion section provides an analysis and interpretation of the findings, compares them with previous studies, identifies limitations, and suggests future directions for research.
This section combines information from the preceding parts of your paper into a coherent story. By this point, the reader already knows why you did your study (introduction), how you did it (methods), and what happened (results). In the discussion, you’ll help the reader connect the ideas from these sections.
Why is it necessary?
The discussion provides context and interpretations for the results. It also answers the questions posed in the introduction. While the results section describes your findings, the discussion explains what they say. This is also where you can describe the impact or implications of your research.
Adds context for your results
Most research studies aim to answer a question, replicate a finding, or address limitations in the literature. These goals are first described in the introduction. However, in the discussion section, the author can refer back to them to explain how the study's objective was achieved.
Shows what your results actually mean and real-world implications
The discussion can also describe the effect of your findings on research or practice. How are your results significant for readers, other researchers, or policymakers?
What to include in your discussion (in the correct order)
A complete and effective discussion section should at least touch on the points described below.
Summary of key findings
The discussion should begin with a brief factual summary of the results. Concisely overview the main results you obtained.
Begin with key findings with supporting evidence
Your results section described a list of findings, but what message do they send when you look at them all together?
Your findings were detailed in the results section, so there’s no need to repeat them here, but do provide at least a few highlights. This will help refresh the reader’s memory and help them focus on the big picture.
Read the first paragraph of the discussion section in this article (PDF) for an example of how to start this part of your paper. Notice how the authors break down their results and follow each description sentence with an explanation of why each finding is relevant.
State clearly and concisely
Following a clear and direct writing style is especially important in the discussion section. After all, this is where you will make some of the most impactful points in your paper. While the results section often contains technical vocabulary, such as statistical terms, the discussion section lets you describe your findings more clearly.
Interpretation of results
Once you’ve given your reader an overview of your results, you need to interpret those results. In other words, what do your results mean? Discuss the findings’ implications and significance in relation to your research question or hypothesis.
Analyze and interpret your findings
Look into your findings and explore what’s behind them or what may have caused them. If your introduction cited theories or studies that could explain your findings, use these sources as a basis to discuss your results.
For example, look at the second paragraph in the discussion section of this article on waggling honey bees. Here, the authors explore their results based on information from the literature.
Unexpected or contradictory results
Sometimes, your findings are not what you expect. Here’s where you describe this and try to find a reason for it. Could it be because of the method you used? Does it have something to do with the variables analyzed? Comparing your methods with those of other similar studies can help with this task.
Context and comparison with previous work
Refer to related studies to place your research in a larger context and the literature. Compare and contrast your findings with existing literature, highlighting similarities, differences, and/or contradictions.
How your work compares or contrasts with previous work
Studies with similar findings to yours can be cited to show the strength of your findings. Information from these studies can also be used to help explain your results. Differences between your findings and others in the literature can also be discussed here.
How to divide this section into subsections
If you have more than one objective in your study or many key findings, you can dedicate a separate section to each of these. Here’s an example of this approach. You can see that the discussion section is divided into topics and even has a separate heading for each of them.
Limitations
Many journals require you to include the limitations of your study in the discussion. Even if they don’t, there are good reasons to mention these in your paper.
Why limitations don’t have a negative connotation
A study’s limitations are points to be improved upon in future research. While some of these may be flaws in your method, many may be due to factors you couldn’t predict.
Examples include time constraints or small sample sizes. Pointing this out will help future researchers avoid or address these issues. This part of the discussion can also include any attempts you have made to reduce the impact of these limitations, as in this study .
How limitations add to a researcher's credibility
Pointing out the limitations of your study demonstrates transparency. It also shows that you know your methods well and can conduct a critical assessment of them.
Implications and significance
The final paragraph of the discussion section should contain the take-home messages for your study. It can also cite the “strong points” of your study, to contrast with the limitations section.
Restate your hypothesis
Remind the reader what your hypothesis was before you conducted the study.
How was it proven or disproven?
Identify your main findings and describe how they relate to your hypothesis.
How your results contribute to the literature
Were you able to answer your research question? Or address a gap in the literature?
Future implications of your research
Describe the impact that your results may have on the topic of study. Your results may show, for instance, that there are still limitations in the literature for future studies to address. There may be a need for studies that extend your findings in a specific way. You also may need additional research to corroborate your findings.
Sample discussion section
This fictitious example covers all the aspects discussed above. Your actual discussion section will probably be much longer, but you can read this to get an idea of everything your discussion should cover.
Our results showed that the presence of cats in a household is associated with higher levels of perceived happiness by its human occupants. These findings support our hypothesis and demonstrate the association between pet ownership and well-being.
The present findings align with those of Bao and Schreer (2016) and Hardie et al. (2023), who observed greater life satisfaction in pet owners relative to non-owners. Although the present study did not directly evaluate life satisfaction, this factor may explain the association between happiness and cat ownership observed in our sample.
Our findings must be interpreted in light of some limitations, such as the focus on cat ownership only rather than pets as a whole. This may limit the generalizability of our results.
Nevertheless, this study had several strengths. These include its strict exclusion criteria and use of a standardized assessment instrument to investigate the relationships between pets and owners. These attributes bolster the accuracy of our results and reduce the influence of confounding factors, increasing the strength of our conclusions. Future studies may examine the factors that mediate the association between pet ownership and happiness to better comprehend this phenomenon.
This brief discussion begins with a quick summary of the results and hypothesis. The next paragraph cites previous research and compares its findings to those of this study. Information from previous studies is also used to help interpret the findings. After discussing the results of the study, some limitations are pointed out. The paper also explains why these limitations may influence the interpretation of results. Then, final conclusions are drawn based on the study, and directions for future research are suggested.
How to make your discussion flow naturally
If you find writing in scientific English challenging, the discussion and conclusions are often the hardest parts of the paper to write. That’s because you’re not just listing up studies, methods, and outcomes. You’re actually expressing your thoughts and interpretations in words.
- How formal should it be?
- What words should you use, or not use?
- How do you meet strict word limits, or make it longer and more informative?
Always give it your best, but sometimes a helping hand can, well, help. Getting a professional edit can help clarify your work’s importance while improving the English used to explain it. When readers know the value of your work, they’ll cite it. We’ll assign your study to an expert editor knowledgeable in your area of research. Their work will clarify your discussion, helping it to tell your story. Find out more about AJE Editing.

Adam Goulston, PsyD, MS, MBA, MISD, ELS
Science Marketing Consultant
See our "Privacy Policy"
Ensure your structure and ideas are consistent and clearly communicated
Pair your Premium Editing with our add-on service Presubmission Review for an overall assessment of your manuscript.
Understanding the Interpretation of Results in Research
Doing the interpretation of results in research is crucial to obtaining valuable findings. Learn how to achieve a good interpretation here!
Research is a powerful tool for gaining insights into the world around us. Whether in academia, industry, or the public sector, research studies can inform decision-making, drive innovation, and improve our understanding of complex phenomena. However, the value of research lies not only in the data collected but also in the interpretation of results. Properly interpreting research findings is critical to extracting meaningful insights, drawing accurate conclusions, and informing future research directions.
In this Mind the Graph article, you’ll understand the basic concept of interpretation of results in research. The article will go over the right procedure for checking, cleaning, and editing your data as well as how to organize it effectively to aid interpretation.
What is the interpretation of results in research?
The process of interpreting and making meaning of data produced in a research study is known as research result interpretation. It entails studying the data’s patterns, trends, and correlations in order to develop reliable findings and make meaningful conclusions.
Interpretation is a crucial step in the research process as it helps researchers to determine the relevance of their results, relate them to existing knowledge, and shape subsequent research goals. A thorough interpretation of results in research may assist guarantee that the findings are legitimate and trustworthy and that they contribute to the development of knowledge in an area of study.
The interpretation of results in research requires multiple steps, including checking, cleaning, and editing data to ensure its accuracy, and properly organizing it in order to simplify interpretation. To examine data and derive reliable findings, researchers must employ suitable statistical methods. They must additionally consider the larger ramifications of their results and how they apply to everyday scenarios.
It’s crucial to keep in mind that coming to precise conclusions while generating meaningful inferences is an iterative process that needs thorough investigation.
The process of checking, cleaning, and editing data

The process of data checking, cleaning, and editing may be separated into three stages: screening, diagnostic, and treatment . Each step has a distinct goal and set of tasks to verify the data’s accuracy and reliability.
Screening phase
The screening process consists of a first inspection of the data to find any errors or anomalies. Running basic descriptive statistics , reviewing data distributions, and discovering missing values may all be part of this. This phase’s goal is to discover any concerns with the data that need to be investigated further.
Diagnostic phase
The diagnostic phase entails a more extensive review of the data to identify particular concerns that must be addressed. Identifying outliers, investigating relationships between variables , and spotting abnormalities in the data are all examples of this. This phase’s goal is to identify any problems with the data and propose suitable treatment options.
Treatment phase
The treatment phase entails taking action to resolve any difficulties found during the diagnostic phase. This may involve eliminating outliers, filling in missing values, transforming data, and editing data. This phase’s goal is to guarantee that the data is reliable, precise, and in the appropriate format for analysis.
Researchers may guarantee that their data is high-quality and acceptable for analysis by using a structured approach to data checking, cleaning, and editing.
How to organize data display and description?
Organizing data display and description is another critical stage in the process of analyzing study results. The format in which data is presented has a significant influence on how quickly it may be comprehended and interpreted. The following are some best practices for data display and description organization.
Best practices for qualitative data include the following:
- Use quotes and anecdotes: Use quotes and anecdotes from participants to illustrate key themes and patterns in the data.
- Group similar responses: Similar replies should be grouped together to find major themes and patterns in the data.
- Use tables: Tables to arrange and summarize major themes, categories, or subcategories revealed by the data.
- Use figures : Figures, such as charts or graphs, may help you visualize data and spot patterns or trends.
- Provide context : Explain the research project’s topic or hypothesis being examined, as well as any important background information, before presenting the findings.
- Use simple and direct language: To describe the data being given, use clear and succinct language.
Best practices for quantitative data include the following:
- Use relevant charts and graphs: Select the right chart or graph for the data being presented. A bar chart, for example, could be ideal for categorical data, but a scatter plot might be appropriate for continuous data.
- Label the axes and include a legend: Label the axes of the chart or graph and include a legend to explain any symbols or colors used. This makes it easier for readers to comprehend the information offered.
- Provide context: Give context to the data that is being given. This may include a brief summary of the research issue or hypothesis under consideration, as well as any pertinent background information.
- Use clear and succinct language: To describe the data being given, use clear and concise language. Avoid using technical jargon or complex language that readers may find difficult to grasp.
- Highlight significant findings: Highlight noteworthy findings in the provided data. Identifying any trends, patterns, or substantial disparities across groups is one example.
- Create a summary table : Provide a summary table that explains the data being provided. Key data such as means, medians, and standard deviations may be included.
3 Tips for interpretation of results in research
Here are some key tips to keep in mind when interpreting research results:
- Keep your research question in mind: The most important piece of advice for interpreting the results is to keep your research question in mind. Your interpretation should be centered on addressing your research question, and all of your analysis should be directed in that direction.
- Consider alternate explanations: It’s critical to think about alternative explanations for your results. Ask yourself whether any other circumstances might be impacting your findings, and carefully assess them. This can assist guarantee that your interpretation is based on the evidence and not on assumptions or biases.
- Contextualize the results: Put the results into perspective by comparing them to past research in the topic at hand. This can assist in identifying trends, patterns, or discrepancies that you may have missed otherwise, as well as providing a foundation for subsequent research.
By following these three tips, you may assist guarantee that your interpretation of data is correct, useful, and relevant to your research topic and the larger context of your field of research.
Professional and custom designs for your publications
Mind the Graph is a sophisticated tool that provides professional and customizable research publication designs. Enhance the visual impact of your research by using eye-catching visuals, charts, and graphs. With Mind the Graph, you can simply generate visually appealing and informative publications that captivate your audience and successfully explain the research’s findings.
Related Articles

Subscribe to our newsletter
Exclusive high quality content about effective visual communication in science.
Sign Up for Free
Try the best infographic maker and promote your research with scientifically-accurate beautiful figures
no credit card required
About Jessica Abbadia
Jessica Abbadia is a lawyer that has been working in Digital Marketing since 2020, improving organic performance for apps and websites in various regions through ASO and SEO. Currently developing scientific and intellectual knowledge for the community's benefit. Jessica is an animal rights activist who enjoys reading and drinking strong coffee.
Content tags
Subgrouping of anxiety symptoms and stress levels in Chinese adolescents: results of a latent profile analysis
- Published: 24 August 2024
Cite this article

- Xuefeng Li 1 ,
- Jingyan Chen 1 ,
- Jinpeng Wang 1 ,
- Jinhong Ding 1 &
- Jing Xiao ORCID: orcid.org/0000-0001-8453-1236 1
The relationship between anxiety symptoms and stress among adolescents has received considerable attention in psychological research. Nonetheless, traditional studies predominantly employ a variable-centered approach, overlooking the potential existence of distinct subgroups within the adolescent population that exhibit unique patterns of anxiety and stress. This study aimed to uncover such subgroupings among Chinese adolescents through latent profile analysis (LPA), utilizing the Multidimensional Anxiety Scale for Children (MASC) and the Adolescent Life Events Questionnaire (ALEQ). The sample comprised 1,027 students aged 14-19 years ( M = 16.26, SD = 0.90) from two middle schools in China. The analysis revealed a three-class solution as the most fitting model, delineating subgroups with Low stress/Low symptoms (51.6%), Low stress/High symptoms (37.2%), and High stress/Low symptoms (11.2%). Subsequent multinomial logistic regression analyses explored the associations between subgroup membership and various demographic and psychological factors, including gender, age, residential background, depression levels, family income, and parents’ marital status and educational levels. The findings indicated a higher prevalence of girls in the “Low stress/High symptom” group and boys in the “High stress/Low symptom” group. Moreover, adolescents from urban areas were more likely to fall into the “High stress/Low symptoms” group, and a strong correlation was observed between higher levels of depression and increased stress or anxiety. These results underscore the importance of incorporating assessments of stress and anxiety into the clinical evaluation of adolescents to enhance the efficacy of psychotherapeutic and preventive interventions.
This is a preview of subscription content, log in via an institution to check access.
Access this article
Subscribe and save.
- Get 10 units per month
- Download Article/Chapter or eBook
- 1 Unit = 1 Article or 1 Chapter
- Cancel anytime
Price includes VAT (Russian Federation)
Instant access to the full article PDF.
Rent this article via DeepDyve
Institutional subscriptions
Data availability
The data that support the findings of this study are available from corresponding author, but restrictions apply to the availability of these data, which were used under licence for the current study and so are not publicly available. The data are, however, available from the authors upon reasonable request.
Anderson, A. S., Siciliano, R. E., Gruhn, M. A., Bettis, A. H., Reising, M. M., Watson, K. H., Dunbar, J. P., & Compas, B. E. (2024). Youth coping and symptoms of anxiety and depression: Associations with age, gender, and peer stress. Current Psychology, 43 (14), 12421–12433. https://doi.org/10.1007/s12144-023-05363-w
Article Google Scholar
Azzi, V., Iskandar, K., Obeid, S., & Hallit, S. (2022). Parental divorce and smoking dependence in Lebanese adolescents: The mediating effect of mental health problems. BMC Pediatrics, 22 (1), 471. https://doi.org/10.1186/s12887-022-03523-8
Article PubMed PubMed Central Google Scholar
Ballesteros, L. M. S., Poleacovschi, C., Weems, C. F., Zambrana, I. G., & Talbot, J. (2023). Evaluating the interaction effects of housing vulnerability and socioeconomic vulnerability on self-perceptions of psychological resilience in Puerto rico. International Journal of Disaster Risk Reduction, 84 , 103476. https://doi.org/10.1016/j.ijdrr.2022.103476
Barbieri, V., Piccoliori, G., Mahlknecht, A., Plagg, B., Ausserhofer, D., Engl, A., & Wiedermann, C. J. (2023). Adolescent mental health during the COVID-19 pandemic: The interplay of age, gender, and mental health outcomes in two consecutive cross-sectional surveys in northern Italy. Behavioral Sciences, 13 (8), 643. https://doi.org/10.3390/bs13080643
Chen, Z. Y., Yang, X. D., & Li, X. Y. (2009). Psychometric features of CES-D in Chinese adolescents. Chinese Journal of Clinical Psychology, 17 (4), 443–445.
Google Scholar
Craig, A., Rochat, T., Naicker, S. N., Mapanga, W., Mtintsilana, A., Dlamini, S. N., Ware, L. J., Du Toit, J., Draper, C. E., Richter, L., & Norris, S. A. (2022). The prevalence of probable depression and probable anxiety, and associations with adverse childhood experiences and socio-demographics: A national survey in south Africa. Frontiers in Public Health, 10 , 986531. https://doi.org/10.3389/fpubh.2022.986531
de la Torre-Luque, A., Borges, G., Benjet, C., Orozco, R., Medina-Mora, M. E., & Ayuso-Mateos, J. L. (2023). Diagnostic profiles in adolescence and emerging adulthood: Transition patterns and risk factors. Spanish Journal of Psychiatry and Mental Health, 16 (1), 42–50. https://doi.org/10.1016/j.rpsm.2022.01.002
Article PubMed Google Scholar
Erskine, H. E., Baxter, A. J., Patton, G., Moffitt, T. E., Patel, V., Whiteford, H. A., & Scott, J. G. (2017). The global coverage of prevalence data for mental disorders in children and adolescents. Epidemiology and Psychiatric Sciences, 26 (4), 395–402. https://doi.org/10.1017/S2045796015001158
Ewing, L., & Hamza, C. A. (2023). A person-centered investigation into the Co-development of perceived stress and internalizing symptoms among post-secondary students. Journal of Youth and Adolescence, 52 (4), 852–865. https://doi.org/10.1007/s10964-023-01738-1
Fassett-Carman, A., Hankin, B. L., & Snyder, H. R. (2019). Appraisals of dependent stressor controllability and severity are associated with depression and anxiety symptoms in youth. Anxiety Stress and Coping, 32 (1), 32–49. https://doi.org/10.1080/10615806.2018.1532504
Goodman, F. R., Birg, J. A., Daniel, K. E., & Kashdan, T. B. (2023). Stress generation in social anxiety and depression: A two-study community assessment. Journal of Affective Disorders, 329 , 285–292. https://doi.org/10.1016/j.jad.2023.02.053
Hankin, B. L., & Abramson, L. Y. (2002). Measuring cognitive vulnerability to depression in adolescence: Reliability, validity, and gender differences. Journal of Clinical Child and Adolescent Psychology, 31 (4), 491–504. https://doi.org/10.1207/S15374424JCCP3104_8
Herbers, J. E., DeCandia, C. J., Volk, K. T., & Unick, G. J. (2023). Profiles and predictors of neurodevelopmental functioning among young children experiencing family homelessness. Early Childhood Research Quarterly, 65 , 407–416. https://doi.org/10.1016/j.ecresq.2023.08.001
Huang, Y., & Chui, H. (2024). Bullying victims’ perceived social support and psychological health and prosocial behavior: A latent profile analysis. Journal of Youth and Adolescence, 53 (7), 1683–1698. https://doi.org/10.1007/s10964-024-01954-3
Li, M., Chen, X., Gong, H., Wang, W., Ji, W., & Liang, S. (2022). Relationship between paternal adult attachment and adolescent anxiety: The chain-mediating effect of paternal psychological flexibility and father-adolescent attachment. International Journal of Psychology, 57 (3), 411–419. https://doi.org/10.1002/ijop.12832
Li, X., Li, W., Li, Q., Ding, J., & Xiao, J. (2023). Residential background as a vulnerability factor for adolescent anxiety in China: A multiwave longitudinal study. International Journal of Stress Management . https://doi.org/10.1037/str0000307
March, J. S., Parker, J. D. A., Sullivan, K., Stallings, P., & Conners, C. K. (1997). The multidimensional anxiety scale for children (MASC): Factor structure, reliability, and validity. Journal of the American Academy of Child and Adolescent Psychiatry, 36 (4), 554–565. https://doi.org/10.1097/00004583-199704000-00019
Mei, K., Zhang, F., Zhang, J., Ming, H., Jiang, Y., & Huang, S. (2024). Perceived social support mitigates the associations among household chaos and health and well-being in rural early adolescents. Journal of Adolescence, 96 (1), 112–123. https://doi.org/10.1002/jad.12260
Merz, E. C., Tottenham, N., & Noble, K. G. (2018). Socioeconomic status, amygdala volume, and internalizing symptoms in children and adolescents. Journal of Clinical Child & Adolescent Psychology, 47 (2), 312–323. https://doi.org/10.1080/15374416.2017.1326122
Nook, E. C., Flournoy, J. C., Rodman, A. M., Mair, P., & McLaughlin, K. A. (2021). High emotion differentiation buffers against internalizing symptoms following exposure to stressful life events in adolescence: An intensive longitudinal study. Clinical Psychological Science, 9 (4), 699–718. https://doi.org/10.1177/2167702620979786
Nurius, P., LaValley, K., & Kim, M.-H. (2020). Victimization, poverty, and resilience resources: Stress process considerations for adolescent mental health. School Mental Health, 12 (1), 124–135. https://doi.org/10.1007/s12310-019-09335-z
Piotrowska, P. J., Stride, C. B., Maughan, B., Ford, T., McIntyre, N. A., & Rowe, R. (2023). Understanding the relationship between family income and conduct problems: Findings from the mental health of children and young people survey. Psychological Medicine, 53 (9), 3987–3994. https://doi.org/10.1017/S0033291722000654
Postigo-Zegarra, S., Schoeps, K., Pérez-Marín, M., Lacomba-Trejo, L., & Valero-Moreno, S. (2024). Personal and family factors for emotional distress in adolescents with chronic disease. Frontiers in Psychology, 14 , 1304683. https://doi.org/10.3389/fpsyg.2023.1304683
Raknes, S., Pallesen, S., Himle, J. A., Bjaastad, J. F., Wergeland, G. J., Hoffart, A., Dyregrov, K., Håland, Å. T., & Haugland, B. S. M. (2017). Quality of life in anxious adolescents. Child and Adolescent Psychiatry and Mental Health, 11 (1), 33. https://doi.org/10.1186/s13034-017-0173-4
Roberts, R. E., & Vernon, S. W. (1983). The center for epidemiologic studies depression scale: Its use in a community sample. American Journal of Psychiatry, 140 (1), 41–46. https://doi.org/10.1176/ajp.140.1.41
Romano, L., Tang, X., Hietajärvi, L., Salmela-Aro, K., & Fiorilli, C. (2020). Students’ trait emotional intelligence and perceived teacher emotional support in preventing burnout: The moderating role of academic anxiety. International Journal of Environmental Research and Public Health, 17 (13), 4771. https://doi.org/10.3390/ijerph17134771
Sánchez Hernández, M. O., Carrasco, M. A., & Holgado-Tello, F. P. (2023). Anxiety and depression symptoms in Spanish children and adolescents: An exploration of comorbidity from the network perspective. Child Psychiatry & Human Development, 54 (3), 736–749. https://doi.org/10.1007/s10578-021-01286-4
Spurk, D., Hirschi, A., Wang, M., Valero, D., & Kauffeld, S. (2020). Latent profile analysis: A review and “how to” guide of its application within vocational behavior research. Journal of Vocational Behavior, 120 , 103445. https://doi.org/10.1016/j.jvb.2020.103445
Tran, B. T., Nguyen, M. T., Nguyen, M. T., Nguyen, T. G., Duc, V. N. H., & Tran, T. T. M. (2023). Mental health and its determinants among adolescents living in families with separated or divorced parents in an urban area of Vietnam. Osong Public Health and Research Perspectives , 14 (4), 300–311. https://doi.org/10.24171/j.phrp.2023.0110
Tsuno, Y. S., & Yamazaki, Y. (2012). Relationships among sense of coherence, resources, and mental health in urban and rural residents in Japan. BMC Public Health, 12 (1), 1107. https://doi.org/10.1186/1471-2458-12-1107
Wang, J., Chen, J., Wang, P., Zhang, S., Li, Q., Lu, S., & Xiao, J. (2024). Identifying internet addiction profiles among adolescents using latent profile analysis: Relations to aggression, depression, and anxiety. Journal of Affective Disorders, 359 , 78–85. https://doi.org/10.1016/j.jad.2024.05.082
Wang, J., Wang, D., Cui, L., McWhinnie, C. M., Wang, L., & Xiao, J. (2017). The “weakest link” as an indicator of cognitive vulnerability differentially predicts symptom dimensions of anxiety in adolescents in China. Journal of Anxiety Disorders, 50 , 69–75. https://doi.org/10.1016/j.janxdis.2017.05.009
Wang, M., Mou, X., Li, T., Zhang, Y., Xie, Y., Tao, S., Wan, Y., Tao, F., & Wu, X. (2023). Association between comorbid anxiety and depression and health risk behaviors among Chinese adolescents: Cross-sectional questionnaire study. JMIR Public Health and Surveillance, 9 , e46289. https://doi.org/10.2196/46289
Wen, L., Yang, K., Cao, Y., Qu, M., & Xiu, M. (2023). Parental marital status and anxiety symptoms in adolescents: The mediating effect of childhood maltreatment. European Archives of Psychiatry and Clinical Neuroscience . https://doi.org/10.1007/s00406-023-01717-4
Whiteside-Mansell, L., McKelvey, L., Saccente, J., & Selig, J. P. (2019). Adverse childhood experiences of urban and rural preschool children in poverty. International Journal of Environmental Research and Public Health, 16 (14), 2623. https://doi.org/10.3390/ijerph16142623
Wu, K., Wang, F., Wang, W., & Li, Y. (2022). Parents’ education anxiety and children’s academic burnout: The role of parental burnout and family function. Frontiers in Psychology, 12 , 764824. https://doi.org/10.3389/fpsyg.2021.764824
Xu, X., Huebner, E. S., & Tian, L. (2021). Co-developmental trajectories of specific anxiety symptoms from middle childhood to early adolescence: Associations with psychological well-being and academic achievement. Journal of Youth and Adolescence, 50 (6), 1140–1156. https://doi.org/10.1007/s10964-021-01411-5
Yalçın, İ, Can, N., Mançe Çalışır, Ö., Yalçın, S., & Çolak, B. (2022). Latent profile analysis of COVID-19 fear, depression, anxiety, stress, mindfulness, and resilience. Current Psychology, 41 (1), 459–469. https://doi.org/10.1007/s12144-021-01667-x
Yao, S., Zou, T., Zhu, X., Abela, J. R. Z., Auerbach, R. P., & Tong, X. (2007). Reliability and validity of the Chinese version of the multidimensional anxiety scale for children among chinese secondary school students. Child Psychiatry and Human Development, 38 (1), 1–16. https://doi.org/10.1007/s10578-006-0039-0
Yarrington, J. S., Metts, A. V., Zinbarg, R. E., Nusslock, R., Wolitzky-Taylor, K., Hammen, C. L., Kelley, N. J., Bookheimer, S., & Craske, M. G. (2023). The role of positive and negative aspects of life events in depressive and anxiety symptoms. Clinical Psychological Science, 11 (5), 910–920. https://doi.org/10.1177/21677026221141654
Zhang, Z., Liao, M., Yao, Z., Hu, B., Xie, Y., Zheng, W., Hu, T., Zhao, Y., Yang, F., Zhang, Y., Su, L., Li, L., Gutknecht, J., & Majoe, D. (2017). Frequency-specific functional connectivity density as an effective biomarker for adolescent generalized anxiety disorder. Frontiers in Human Neuroscience, 11 , 549. https://doi.org/10.3389/fnhum.2017.00549
Zhou, Z., Li, Y., Zhang, Y., Liu, J., Ai, H., Liu, M., Qiu, J., Luo, Y., & Xu, P. (2023). Differential effects of generalized anxiety and separation anxiety on brain structural development during adolescence. Journal of Affective Disorders, 339 , 478–485. https://doi.org/10.1016/j.jad.2023.07.056
Download references
Acknowledgements
The authors thank all those who provided support and all participants for attending the study.
The authors did not receive support from any organization for the submitted work.
Author information
Authors and affiliations.
Beijing Key Laboratory of Learning and Cognition and Department of Psychology, Capital Normal University, Beijing, 100048, China
Xuefeng Li, Jingyan Chen, Jinpeng Wang, Jinhong Ding & Jing Xiao
You can also search for this author in PubMed Google Scholar
Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by [Xuefeng Li], [Jingyan Chen] and [Jinpeng Wang]. The first draft of the manuscript was written by [Xuefeng Li] and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Correspondence to Jinhong Ding or Jing Xiao .
Ethics declarations
Ethics approval.
Approval was obtained from the ethics committee of Psychology Committee of Capital Normal University. The procedures used in this study adhere to the tenets of the Declaration of Helsinki.
Consent to participate
Informed consent was obtained from all individual participants included in the study.
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Publisher's note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Reprints and permissions
About this article
Li, X., Chen, J., Wang, J. et al. Subgrouping of anxiety symptoms and stress levels in Chinese adolescents: results of a latent profile analysis. Curr Psychol (2024). https://doi.org/10.1007/s12144-024-06423-5
Download citation
Accepted : 12 July 2024
Published : 24 August 2024
DOI : https://doi.org/10.1007/s12144-024-06423-5
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Adolescents
- Anxiety symptoms
- Stress levels
- Latent profile analysis
- Find a journal
- Publish with us
- Track your research
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- Open access
- Published: 23 August 2024
Macrophage ILF3 promotes abdominal aortic aneurysm by inducing inflammatory imbalance in male mice
- Zhao-yang Wang ORCID: orcid.org/0000-0003-0898-4801 1 , 2 na1 ,
- Jie Cheng 1 na1 ,
- Ying Wang 1 na1 ,
- Hai-tao Yuan 2 na1 ,
- Shao-jie Bi 3 na1 ,
- Shuang-xi Wang ORCID: orcid.org/0000-0003-3275-7916 1 ,
- Ya-min Hou 1 ,
- Xu Zhang 1 ,
- Bo-han Xu 1 ,
- Ze-ying Wang 1 ,
- Yun Zhang ORCID: orcid.org/0000-0003-4432-6144 1 ,
- Wen-jian Jiang 4 ,
- Yu-guo Chen ORCID: orcid.org/0000-0001-9501-2546 5 &
- Ming-xiang Zhang 1
Nature Communications volume 15 , Article number: 7249 ( 2024 ) Cite this article
1 Altmetric
Metrics details
- Chronic inflammation
Imbalance of proinflammatory and anti-inflammatory responses plays a crucial role in the progression of abdominal aortic aneurysms. ILF3, a known modulator of the innate immune response, is involved in cardiovascular diseases. This study aims to investigate the role of ILF3 in abdominal aortic aneurysm formation. Here, we use multi-omics analyzes, transgenic male mice, and multiplex immunohistochemistry to unravel the underlying involvement of ILF3 in abdominal aortic aneurysms. The results show that macrophage ILF3 deficiency attenuates abdominal aortic aneurysm progression, while elevated macrophage ILF3 exacerbates abdominal aortic aneurysm lesions. Mechanistically, we reveal that macrophagic ILF3 increases NF-κB activity by hastening the decay of p105 mRNA, leading to amplified inflammation in macrophages. Meanwhile, ILF3 represses the anti-inflammatory action by inhibiting the Keap1-Nrf2 signaling pathway through facilitating the ILF3/eIF4A1 complex-mediated enhancement of Keap1 translational efficiency. Moreover, Bardoxolone Methyl treatment alleviates the severity of abdominal aortic aneurysm lesions in the context of elevated ILF3 expression. Together, our findings underscore the significance of macrophage ILF3 in abdominal aortic aneurysm development and suggest its potential as a promising therapeutic target for abdominal aortic aneurysms.
Similar content being viewed by others
Loss of myeloid Tsc2 predisposes to angiotensin II-induced aortic aneurysm formation in mice

JAK2V617F mutation drives vascular resident macrophages toward a pathogenic phenotype and promotes dissecting aortic aneurysm
Administration of anti-inflammatory M2 macrophages suppresses progression of angiotensin II-induced aortic aneurysm in mice
Introduction.
Abdominal aortic aneurysm (AAA) is a complex fatal vascular disease characterized by irreversible enlargement of the aortic diameter, accompanied by focal inflammation, turbulent proteolytic activities, and extracellular matrix (ECM) degradation 1 , 2 , 3 . Inflammatory cell infiltration and intense inflammatory response play crucial roles in AAA initiation and progression 1 , 4 . Recently, there has been an emphasis on the role of immune regulation in maintaining the inflammatory balance during AAA progression 5 . Clinical investigations have found that aneurysm size in patients with AAA is dependent on the imbalance of pro-inflammatory and anti-inflammatory responses 6 . Moreover, serum levels of the pro-inflammatory cytokine interleukin 6 (IL-6) were significantly increased in AAA patients, in contrast to a notable reduction in the anti-inflammatory cytokine Interleukin-10 (IL-10) 2 . Similar findings have been observed in human AAA explant cultures and murine models 5 , 7 . Recent microarray-based gene expression studies have confirmed that AAA is a chronic inflammatory disease, with the actions of activated macrophage subsets playing a significant role 7 . Macrophage infiltration into the adventitia serves as a seminal event in the onset of AAA pathogenesis, which mediates inflammatory homeostasis by producing chemokines and cytokines that is critical to aneurysmal progression 8 , 9 . However, the specific molecular mechanism of macrophage infiltration and inflammation in aneurysm dilatation are not yet fully delineated. Besides, no explicit therapy targeting the immunoinflammatory response is currently available to impede excessive inflammatory response and remodeling of the abdominal aortic wall.
Interleukin enhancer-binding factor 3 (ILF3) is an RNA-binding protein initially identified as a positive regulator of IL2 gene expression 10 . Subsequent reports have indicated that ILF3 is also involved in dendritic cell maturation, innate immune response, and carcinoma progression 11 , 12 . More broadly, there is evidence that ILF3 is associated with cardiovascular diseases, such as stroke and myocardial infarction 13 , 14 . It has been reported that ILF3 is involved in the enhanced vascular smooth muscle cell apoptosis due to the overexpression of circACTA2 via reducing CDK4 mRNA stability 15 . Nevertheless, no studies have been conducted to explore the role of ILF3 in AAA.
The current study identified ILF3 as the target gene because of its high expression and specificity with macrophages in AAA lesions, as determined through bioinformatic analyzes and immunoassays. By employing conditional macrophage ILF3-deficient and overexpression mouse strains, we revealed the detrimental role of macrophage ILF3 in AAA development, with macrophage ILF3 being positively correlated to the intensity of focal vascular inflammatory response. Mechanistic studies further demonstrated that macrophage ILF3 acts as a dual modulator in the inflammatory cascade, where it both accelerates p105 mRNA instability to potentiate NF-κB-driven pro-inflammatory cascades and enhances Keap1 translational efficiency via the ILF3/eIF4A1 complex to dampen the Keap1-Nrf2-mediated anti-inflammatory responses. Furthermore, interventions targeting ILF3 gene expression, as well as pharmacological interventions using BM, were found to effectively mitigate the exacerbated progression of AAA caused by high ILF3 expression. These findings thus help elucidate the mechanisms of aortic aneurysm formation and provide a framework to guide the future development of treatments for patients with AAA by targeting ILF3.
ILF3 is elevated in human and murine AAA tissues
To pinpoint the candidate genes potentially associated with AAA, we downloaded publicly available data from the Gene Expression Omnibus (GEO) database (GSE57691 and GSE12591), and DEGs were acquired via GEO2R analysis. 715 genes displayed the same trend in human and mouse AAA samples compared with normal controls, among which 410 genes were downregulated and 305 were upregulated (Fig. 1A ). Among the top differentially expressed genes shown in the heatmap, ILF3 expression was significantly elevated in patients with AAA (Fig. 1B , and Supplementary Fig. 1A ) and Ang-II-induced AAA mice (Fig. 1C ), indicating a vital role of ILF3 in AAA progression. Besides, western blot (Figs. 1D, E ), quantitative PCR (Fig. 1F ), and immunohistochemical staining results (Supplementary Fig. 1B ) showed that the expression of ILF3 was augmented in human AAA (patients’ information in Supplementary Table 1 ). Similar results were also seen in Ang-II-induced mouse AAA compared with the control group (Figs. 1G–I ; and Supplementary Fig. 1C ).

A Venn diagram showing the overlap of differentially expressed genes (DEGs) between human and mouse samples. B Heatmap showing gene expression in human control and AAA aorta tissues ( p < 0.05, Foldchange>1.2). C Heatmap showing gene expression in mouse control and angiotensin II–induced AAA aorta tissues ( p < 0.05, Foldchange>1.2). D and E Western blot analysis of ILF3 levels in aortas of controls and patients with AAA ( n = 10 per group). F Relative mRNA levels of ILF3 in aorta tissues from controls and patients with AAA ( n = 10 per group). The control sample was set as 1. G – H Western blot analysis of ILF3 levels in aortas from mouse control and angiotensin II–induced AAA samples ( n = 7 per group). I RT-PCR of ILF3 mRNA in aorta tissues from control and angiotensin II–induced AAA mouse samples ( n = 7 per group). The control sample was set as 1. J ILF3 + macrophages from human AAA and non-aneurysmal samples were projected onto the t-SNE plot. The color intensity of each dot corresponds to the average gene expression across all cells. K Dot plot representing the ILF3 expression of macrophage clusters in human AAA and non-aneurysmal samples. The color of the dot represents the ILF3 expression level (average expression), and the size of the dot corresponds to the percentage of cells expressing ILF3 (percent expressed) across the human AAA and non-aneurysmal samples. L FACS analysis quantified the proportion of ILF3 + macrophages within lesions of ApoE −/− mice subjected to Ang II infusion at specified time ( n = 4 per group). M Representative confocal images of ILF3 (green) colocalized with CD68 (a marker of macrophage, red) in aorta tissues from controls and AAA lesions of humans. Nuclei were stained with DAPI (blue), and yellow represents merged images (Scale bar=25 μm; n = 8 per group). N Representative confocal images of ILF3 (green) colocalized with CD68 (marker of macrophage, red) in aorta tissues from ApoE −/− mice treated with Ang II for different intervals (Scale bar=25 μm; n = 8 per group). Data are represented as mean ± SD. For comparisons between two groups, a Student’s t-test with two-tailed analysis (1 B –1 C , 1 E –1 F , 1 H –1 I ). For comparisons between more than two groups, one-way ANOVA followed by Tukey’s post hoc multiple comparisons test was used (1 L ). Boxplots were created using the first and third quartiles to define the bounds of the box. The minima are defined as the first quartile minus 1.5 times the inter-quartile range (IQR), and the maxima are defined as the third quartile plus 1.5 times the IQR. Outliers are displayed as individual points (1 E –1 F , 1 H –1 I and 1 L ).
Macrophage ILF3 is augmented during AAA progression
To discern ILF3’s spatial role in AAA lesions, we utilized Seurat for analyzing human single-cell RNA sequencing data (GSE166676) from the GEO database. Through UMAP and canonical markers, we classified cells into seven types (Supplementary Figs. 1D, E ); the representative genes in each cell type are shown in Supplementary Fig. 1F . GO analysis showed that the immune response, particularly mononuclear cell activation and focal adhesion, were elevated in ILF3 + cells compared with ILF3 − clusters (Supplementary Fig. 1G ). Furthermore, a substantial increase in ILF3-positive macrophages was observed in human AAA lesions (Figs. 1J, K ; Supplementary Figs. 1H–K ), underscoring the critical role of macrophage ILF3 in AAA progression. To further explore ILF3’s function in macrophages within AAA, we established AAA models at different pathological stages using Ang II induction over varied timeframes. Flow cytometry demonstrated a progressive rise in ILF3-positive macrophages with prolonged Ang II exposure (Fig. 1L ; Supplementary Fig. 2A ). Additionally, increased ILF3 expression in both human and mouse AAA samples was verified, with clear macrophage colocalization noted in immunofluorescence assays (Figs. 1M, N ). Notably, ILF3 protein levels in bone marrow-derived and peritoneal macrophages from wild-type C57BL/6 J mice showed a time and dose-dependent increase upon Ang II treatment (Supplementary Figs. 2B–E ). Collectively, these findings compellingly suggest that the marked elevation of ILF3 in macrophages plays a crucial role in AAA development.
Macrophage ILF3 overexpression exacerbates AAA formation
Observing elevated ILF3 levels in AAA, particularly in macrophages, we investigated macrophage ILF3 overexpression’s impact on AAA using ILF3 conditional overexpression mice (ILF3 Flox/Flox LysMCre + ; hereafter referred to as ILF3 M-Tg ) and its littermates (ILF3 Flox/Flox LysMCre - ; hereafter referred to as ILF3 M-WT ) in an ApoE −/− background (Supplementary Figs. 3A – 3D ). Following a 4-week saline infusion, no aneurysms were observed in either group (Fig. 2A ). However, under Ang II induction, both groups experienced a significant rise in systolic blood pressure with Ang II, with no differences between the ILF3 M-Tg ApoE −/− and ILF3 M-WT ApoE −/− mice (Supplementary Table 2 ). Notably, ILF3 M-Tg ApoE −/− mice displayed more pronounced aortic dilation compared with ILF3 M-WT ApoE −/− mice (Fig. 2A ). The incidence of AAA formation (Fig. 2B ) and the rupture rate of AAA (Fig. 2C ) in ILF3 M-Tg ApoE −/− mice was markedly higher than that of ILF3 M-WT ApoE −/− group through the 4-week Ang II exposure. Among those subjected to Ang II infusion, 68.75% (22/32) of ILF3 M-Tg ApoE −/− mice died, apparently higher than the mortality rate of ILF3 M-WT ApoE −/− (37.50%, 12/32; Fig. 2D ). Vascular ultrasound imaging and maximal external aortic diameter detection further corroborated these findings, revealing more pronounced dilation in the ILF3 M-Tg ApoE −/− mice post-Ang II infusion (Fig. 2E ; Supplementary Fig. 3E ). Histological analysis indicated no elastin disruption under baseline conditions; however, ILF3 overexpression in macrophages significantly exacerbated elastin degradation in Ang II-induced AAA models (Figs. 2F, G ).

The 8-week-old male ILF3 M-Tg ApoE −/− and ILF3 M-WT ApoE −/− mice were infused with Ang ll or saline (sham group). A Schematic representation of study design and representative images showing the morphology of the whole aorta from all groups showing the macroscopic characteristics of aneurysms. B – D The AAA incidence ( B ) rupture rate ( C ) and Kaplan-Meir survival curve ( D ) across different groups (sham group, n = 6; Ang II group, n = 32). Statistical analyses employed a chi-square test with a two-tailed analysis for ( B , C ) and the Log-rank test for ( D ). E Representative ultrasound images of aorta across different groups (sham group, n = 6; Ang II group, n = 12). F and G Representative hematoxylin and eosin (HE) and Verhoeff-Van Gieson (VVG) staining ( F ) and elastin degradation grading ( G ). (sham group, n = 5; Ang II group, n = 12; Scale bar=100 μm). 8-week-old male ILF3 M-Tg and ILF3 M-WT mice were infused with CaCl 2 or saline (sham group). H and I Schematic representation of study design and representative photographs (H) of saline-treated and CaCl 2 -induced aneurysm and maximal abdominal aortic diameter (I; sham group, n = 6; Cacl 2 group, n = 12). Data are represented as mean ± SD. For comparisons between two groups, an unpaired Student’s t-test with two-tailed analysis (2 G and 2 I ). Boxplots were created using the first and third quartiles to define the bounds of the box, with the median shown in a circle. The minima are defined as the first quartile minus 1.5 times the inter-quartile range (IQR), and the maxima are defined as the third quartile plus 1.5 times the IQR. Violin plots display the density distribution of the data points through smoothed histograms.
To further validate the role of macrophage ILF3 in AAA, we evaluated the effects of macrophage ILF3 overexpression in CaCl 2 -induced AAA model mice. Consistent with the results observed in Ang-II-induced AAA model mice, ILF3 M-Tg mice were more susceptible to abdominal aortic dilation with increased maximal abdominal aortic diameter in CaCl 2 -induced AAA model (Figs. 2H, I ). Moreover, HE and VVG staining showed that elastic lamellae of ILF3 M-Tg mice were disrupted and degraded to a greater extent than that of ILF3 M-WT mice (Supplementary Figs. 3F, G ). Collectively, these results underscore the pivotal role of macrophage-derived ILF3 in promoting AAA formation and progression in both CaCl 2 - and Ang II-induced models.
Macrophage ILF3 deficiency attenuates AAA formation
To identify whether macrophage ILF3 deficiency is protective against AAA formation, we constructed macrophage-ILF3 knockout mice (ILF3 flox/flox LysMCre + ; hereafter referred to as ILF3 M-KO ) in ApoE −/− mice (Supplementary Figs. 4A, B ). Upon four weeks of Ang II or saline infusion, ILF3 M-KO ApoE −/− mice exhibited reduced aortic dilation in response to Ang II, while saline infusion had no discernible effect on either group (Fig. 3A ). Importantly, blood pressure measurements remained consistent between the two genotypes, both pre- and post-Ang II infusion (Supplementary Table 3 ). Upon Ang II treatment, ILF3 M-KO ApoE −/− mice showed a decreased incidence of AAA (Fig. 3B ) and a reduced rupture rate (Fig. 3C ) compared to ILF3 M-WT ApoE −/− mice. And the mortality rate of ILF3 M-WT ApoE −/− mice post-Ang II infusion was 40.63% (13/32), evidently higher than the 15.79% (6/38) observed in ILF3 M-KO ApoE −/− (Fig. 3D ). Moreover, after Ang II infusion, ILF3 M-KO ApoE −/− mice exhibited a discernible reduction in aortic dilation relative to ILF3 M-WT ApoE −/− mice (Supplementary Fig. 4C ; Fig. 3E ). Histological assessments using HE and VVG staining revealed that ILF3-deficient mice presented with moderate elastic degradation (Fig. 3F, G ). Using CaCl 2 -induced AAA model, we also found that ILF3 M-KO mice were less susceptible to abdominal aortic dilation with mitigated elastic degradation (Figs. 3H, I ; Supplementary Figs. 4D, E ). Overall, these results showed that macrophage ILF3 deficiency attenuated AAA formation, thereby highlighting its promise as a therapeutic target for AAA management.

The 8-week-old male ILF3 M-KO ApoE −/− and ILF3 M-WT ApoE −/− mice were infused with Ang ll or saline (sham group). A Schematic representation of study design and representative images showing the morphology of the whole aorta from all groups showing the macroscopic characteristics of aneurysms. B – D The AAA incidence ( B ) rupture rate ( C ) and Kaplan-Meir survival curve ( D ) across different groups (sham group, n = 6; ILF3 M-WT ApoE −/− + Ang II group, n = 32; ILF3 M-KO ApoE −/− + Ang Ii, n = 38). Statistical analyses employed a chi-square test with a two-tailed analysis for ( B , C ) and the Log-rank test for ( D ). E Representative ultrasound images of aorta across different groups (sham group, n = 6; Ang II group, n = 12). F and G Representative hematoxylin and eosin (HE) and Verhoeff-Van Gieson (VVG) staining ( F ) and elastin degradation grading ( G ). (sham group, n = 5; Ang II group, n = 12; Scale bar=100 μm). 8-week-old male ILF3 M-KO and ILF3 M-WT mice were infused with CaCl 2 or saline (sham group). H and I Schematic representation of study design and representative photographs ( H ) of saline-treated and CaCl 2 -induced aneurysm and maximal abdominal aortic diameter ( I ) sham group, n = 6; CaCl 2 group, n = 12). Data are represented as mean ± SD. For comparisons between two groups, an unpaired Student’s t-test with two-tailed analysis (3 G and 3 I ). Boxplots were created using the first and third quartiles to define the bounds of the box, with the median shown in a circle. The minima are defined as the first quartile minus 1.5 times the inter-quartile range (IQR), and the maxima are defined as the third quartile plus 1.5 times the IQR. Violin plots display the density distribution of the data points through smoothed histograms.
Macrophage ILF3 contributes to Ang-II-induced inflammatory cell infiltration and the inflammatory response
To uncover the mechanism underlying the potential role of macrophage-derived ILF3 on AAA, we analyzed the enrichment results based on the scRNA-seq data 16 . As shown by the Gene Ontology (GO, Fig. 4A ) and KEGG analysis (Fig. 4B ), macrophage ILF3 was significantly associated with immune response and pathways related to immune response (such as NF-κB, TNF, and HIF1 signaling pathway were apparently enriched with macrophage ILF3). Macrophages play a pivotal role in AAA pathogenesis, where they mainly accumulate in the adventitial layer, serving as essential producers and responsive entities to inflammatory mediators, thereby orchestrating the intricate inflammatory dynamics within the aortic wall 17 .

A Representative Gene Ontology terms of differentially expressed genes (DEGs) enriched in ILF3 + macrophage clusters based on functional enrichment analysis. B Representative Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway data for macrophage DEGs enriched in ILF3 + vs ILF3 - . C Immuno-staining of macrophage-derived MCP-1, TNF-α, IL-6, and IL-10 levels in AAA lesions of ILF3 M-WT ApoE −/− and ILF3 M-KO ApoE −/− mice treated with Ang II for four weeks ( n = 6 per group; Scale bar=25 μm). D Plasma IL-6, TNF-α, MCP-1, IL10 levels in ILF3 M-WT ApoE −/− and ILF3 M-KO ApoE −/− mice treated with Ang II for four weeks ( n = 10 per group). E Western blot analysis and quantification of ILF3, MCP1, TNF-α, IL-6, and IL-10 expression in peritoneal macrophages from the ILF3 deficiency and control groups with or without Ang II treatment (1 μM; n = 6 per group). Data are represented as mean ± SD. P values were calculated by one-sided Fisher’s exact test in (4 A and 4 B ). For comparisons between two groups, an unpaired Student’s t-test with two-tailed analysis was performed if the data met the assumptions of normality and equal variances; otherwise, the Wilcoxon rank-sum test was used (4 D ). For comparisons among four groups, one-way ANOVA followed by Tukey’s post hoc multiple comparisons test was used (4 E ).
Utilizing immunofluorescent double staining techniques, we found the number of CD68 + macrophages was reduced in ILF3 M-KO ApoE −/− mice (Fig. 4C ) and increased in ILF3 M-Tg ApoE −/− mice compared with control mice (Supplementary Fig. 5A ). Meanwhile, the results revealed the expression of several inflammatory cytokines and chemokines, including monocyte chemoattractant protein-1 (MCP-1), IL-6, TNF-α, and IL-10 exhibited a pronounced decrement in the ILF3 M-KO ApoE −/− mice (Fig. 4C ). Conversely, these were significantly upregulated in the ILF3 M-Tg ApoE −/− group (Supplementary Fig. 5A ). Notably, the macrophage-produced immunosuppressive cytokine IL-10 expression was diametrically opposed to the aforementioned pro-inflammatory mediators (Fig. 4C ; Supplementary Fig. 5A ). Moreover, similar results were further validated in the AAA mouse model by assessing serum samples through Elisa (Fig. 4D ; Supplementary Fig. 5B ).
In vitro, we treated the isolated peritoneal macrophages (Fig. 4E ) and bone marrow-derived macrophages (Supplementary Fig. 5C ) from the macrophage ILF3 deficiency and wild-type mice with Ang II. Immunoblotting analysis demonstrated that macrophages deficient in ILF3 showed decreased expression of MCP-1 and inflammatory cytokines (IL-6 and TNF-α). IL-10 levels were, however, intensified due to the lack of ILF3. The above results showed that deficiency of macrophage-derived ILF3 attenuates Ang-II-induced inflammatory cell infiltration and concomitant inflammation, while an elevation in ILF3 expression exerts the opposite effect.
Macrophage ILF3 mediates NF-κB and Keap1-Nrf2 pathways via p105 and Keap1
To investigate the machinery between ILF3 and subsequent physio-biologic features of macrophages, we conducted proteomics using peritoneal macrophages from mice of ILF3 M-WT and ILF3 M-KO . Our findings revealed 1233 uniquely differentially expressed proteins attributable to ILF3 deficiency (Supplementary Fig. 6A ; and Supplementary Data 1 ), although a small amount of residual expression of ILF3 was observed after its knockout. GO enrichment analysis, aligning with our scRNA-seq results, demonstrated ILF3’s strong association with inflammatory response (Supplementary Fig. 6B ). KEGG pathway analysis highlighted ILF3’s role in key pathways such as Fluid Shear Stress and Atherosclerosis and NF-κB pathway (Fig. 5A ), with significant alterations in p105 (nfkb1) and Keap1 (Fig. 5B ). These factors are known to regulate various inflammatory cytokines and chemokines via NF-κB and Keap1-Nrf2 signaling pathway 18 , 19 , 20 . Western blotting validated these findings, showing decreased Keap1 and increased p105 and p50 in ILF3 M-KO mice across both peritoneal and bone marrow-derived macrophages (Supplementary Figs. 6C – 6F ). Additionally, ILF3 deficiency could inhibit the Ang II-induced p65 nuclear translocation and increased cytoplasmic expression of p105 and p50 (Fig. 5C–E ; Supplementary Figs. 6G, H ), and moderated impaired Nrf2 nuclear translocation while lowering Keap1 expression (Figs. 5C, D, F ; and Supplementary Fig. 6G, H ). Furthermore, Co-IP analysis showed more prevalence of p50:p105 and p50:p50 complexes, while less p50:p65 complex in ILF3 M-KO peritoneal macrophages (Figs. 5G, H ). Given that p65 and Nrf2 are well-established transcription factors in the inflammatory response, Electrophoretic Mobility Shift Assay (EMSA) showed reduced NF-κB DNA binding in ILF3-deficient macrophages (Fig. 5I ). On the contrary, ILF3 deficiency increased Nrf2-ARE DNA binding, especially, the Nrf2 binding to the ARE sequence of key inflammatory (e.g., IL-6, MCP1; Fig. 5J , Supplementary Figs. 6I, J ). Collectively, the above results indicate that ILF3 mediates the inflammatory response via the NF-κB and Keap1-Nrf2 signaling.

A Bubble plot showing Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of differentially expressed genes (DEGs) in macrophages of ILF3 M-WT vs. ILF3 M-KO mice (one-sided Fisher’s exact test). The top 10 significantly enriched KEGG pathways. B KEGG pathway analysis for DEGs in significant KEGG pathway terms was visualized as a chord dendrogram composed of ribbons. C – D Western blot analysis and quantification of cytoplasmic and nuclear p105, p50, p65, Keap1, and Nrf2 expression in peritoneal macrophages of ILF3 deficiency and control groups with or without Ang II treatment (1 μM; n = 6 per group). E – F Immunofluorescence staining for p65 (red, E ) and Nrf2 (green, F ) in peritoneal macrophages from ILF3 deficiency and control mice with or without Ang II treatment (1 μM); nuclei were stained with DAPI (blue, Scale bar, 20 μm; n = 6 per group). G – H Co-immunoprecipitation (Co-IP) assay and visualization of interactions between p50, p105, and p65 with an anti-p50 monoclonal antibody in peritoneal macrophages from ILF3 deficiency and control mice after Ang II stimulus (1μm, 24 h, n = 3 per group). I – J , EMSA analysis showing the DNA binding activity of both NF-κ B ( I ) and Nrf2 ( J ) in nuclear extracts of peritoneal macrophages from ILF3 deficiency and control mice after Ang II stimulus (1 μm, 24 h, n = 6 per group). K – L Western blot analysis and quantification of MCP-1, TNF-α, IL-6, and IL-10 expression in peritoneal macrophages from ILF3 M-WT and ILF3 M-Tg mice treated with si-Keap1 and AdV-p105 inhibitor under the stimulus without or with Ang II (1 μM; n = 5 per group). Data are represented as mean ± SD. For comparisons between more than two groups, one-way ANOVA followed by Tukey’s post hoc multiple comparisons test was used (5 D , 5 L ). For comparisons between two groups, an unpaired Student’s t-test with two-tailed analysis (5 H ).
To elucidate whether ILF3’s regulation of NF-κB and Keap1-Nrf2 signaling depends on p105 and Keap1 expression, we used p105-overexpressing adenovirus (AdV-p105, Supplementary Figs. 7A–B ) and Keap1 small interfering RNA (si-Keap1, Supplementary Figs. 7C–D ). Western blotting and immunofluorescence revealed that p105 overexpression limited p65 translocation in ILF3 M-Tg mice, without altering Keap1/Nrf2 expression (Supplementary Fig. 8A–D ). EMSA comfirmed that ectopic p105 expression reduced NF-κB activation, leaving the Nrf2 pathway unaffected caused by overexpression of macrophage ILF3 (Supplementary Figs. 8E, F ). Additionally, Keap1 knockdown restored nuclear Nrf2 expression (Supplementary Figs. 8C, D ; Supplementary Figs. 8G, H ) and Nrf2 activation (Supplementary Fig. 8I-K ) under ILF3 overexpression, without inhibiting NF-κB pathway (Supplementary Fig. 8L ). And both si-Keap1 and AdV-p105 significantly reduced ILF3 overexpression-induced inflammation in macrophages (Figs. 5K, L ; Supplementary Fig. 9 ). In summary, our data demonstrate that ILF3 deficiency modulates inflammatory balance through dual mechanisms: inhibiting NF-κB via p105 upregulation and activating Keap1-Nrf2 pathway due to reduced Keap1 expression.
Macrophage ILF3 promotes p105 mRNA instability and Keap1 translation efficiency
To unravel the underlying mechanisms behind the altered expression of Keap1 and p105 in ILF3-deficient macrophages, we initially assessed protein degradation pathways as contributing factors. This was achieved by employing MG132, a proteasome inhibitor, and hydroxychloroquine (HCQ), a classical lysosomal inhibitor. Neither of these inhibitors altered the changes observed in peritoneal macrophages (Supplementary Fig. 10A ) and bone marrow-derived macrophages (Supplementary Fig. 10B ) from ILF3 M-KO mice. Considering ILF3’s established role in RNA metabolism, a fact substantiated by both our enrichment analysis and previous studies 10 , we next conducted RNA Immunoprecipitation sequencing (RIP-seq). The results revealed that 83.2% of ILF3-bound RNAs were involved in protein coding, highlighting ILF3’s critical role in biological functions via RNA metabolism (Fig. 6A ). Furthermore, RIP-seq showed ILF3 was able to bind to both p105 and Keap1 RNA (Figs. 6B, C ; Supplementary Fig. 10C, D ). RT-PCR analysis indicated that ILF3 deficiency increased p105 mRNA expression without affecting Keap1 RNA levels in peritoneal and bone marrow-derived macrophages (Fig. 6D ; Supplementary Fig. 10E ). Assessment of mRNA stability showed that deficiency of ILF3 slowed the degradation of p105 mRNA but had no impact on Keap1 mRNA (Fig. 6E ; Supplementary Figs. 10F–H ). To further explore ILF3’s influence on Keap1 protein expression, we performed IP-MS and found no interaction between ILF3 and Keap1. However, eIF4A1 emerged as a potential mediator, given its role in initiating cap-dependent eukaryotic protein translation (Fig. 6F ). Subsequent Co-IP confirmed the interaction between ILF3 and eIF4A1 (Fig. 6G, H ). Moreover, we employed the catRAPID method to estimate the binding potential of eIF4A1 protein to Keap1 mRNA 21 . The catRAPID identified a direct interaction between eIF4A1 protein and Keap1 mRNA, with a Global Score of 1.00 (scaling from 0 to 1, where 0 indicates no RNA-binding ability and 1 indicates strong affinity). And it revealed that the predominant binding site for eIF4A1 is located in the 5’ UTR of Keap1 mRNA (Supplementary Fig. 10I ), which was further validated by an in vitro RIP assay (Fig. 6I ). These findings propose the assembly of an ILF3/eIF4A1/Keap1 mRNA complex, which might account for the observed reduction in Keap1 protein levels. Ribosomal profiling confirmed our suspicions, and similar results were acquired using RT-PCR, which showed that macrophage ILF3 deficiency inhibits the translation efficiency of Keap1 (Figs. 6J, K ; and Supplementary Data 2 ). Besides, results showed knockdown of eIF4A1 via si-eIF4A1 suppressed the Keap1 translation efficiency (Supplementary Fig. 10J ) and inhibited the elevated translation of Keap1 due to ILF3 overexpression (Supplementary Fig. 10K ). In summary, our findings prove that ILF3 deficiency in macrophages enhances p105 mRNA stability and inhibits Keap1 translation, thereby modulating the inflammatory response through the NF-κB and Keap1-Nrf2 pathways.

A Distribution of ILF3-binding regions on the genome relative to RIP-Sequence. B – C Representative ILF3 RIP-seq peaks were shown as track signals in an integrative genomic viewer of p105 (Nfkb1) ( B ) and Keap1 ( C ). Peak area is highlighted in light blue and peak region is denoted as red rectangle. D Relative mRNA levels of Keap1 and p105 in peritoneal macrophages from ILF3 M-WT and ILF3 M-KO mice ( n = 6 per group). E Relative p105 remaining RNA levels at different time points after Actinomycin-D (Act-D) treatment in peritoneal macrophages from ILF3 M-WT and ILF3 M-KO mice ( n = 6 per group). F , Venn diagrams showing the proteins immunoprecipitated with anti-IgG and anti-ILF3 antibodies in wild-type peritoneal macrophages. Blue represents proteins immunoprecipitated with anti-ILF3 under basal conditions (NC). Pink represents proteins immunoprecipitated with anti-IgG. G – H , Co-IP assay showing interactions between ILF3 and eIF4A1 with an anti-ILF3 antibody ( G ) and anti-eIF4A1 ( H ) antibody in peritoneal macrophages from control mice ( n = 3 per group). I RIP assay for enrichment of Keap1 mRNA with eIF4A1 in wild-type peritoneal macrophages; IgG served as an internal control ( n = 3). J Translation profiles for Keap1 computed from Ribo-seq data without induction. Positions of the genetic parts and gene are shown above the profiles. The Y axis indicates sequence depth. K, Box plot showing the translation efficiency (TE) of Keap1 mRNAs in peritoneal macrophages from ILF3 M-WT and ILF3 M-KO mice ( n = 3 per group). Data are represented as mean ± SD. For comparisons between two groups, an unpaired Student’s t-test with two-tailed analysis (6D and 6 K). For 6E, statistical analysis was performed using Two-way ANOVA and Sidak’s multiple comparison post-hoc test (*** p < 0.001).
Pharmacological therapy with BM alleviates macrophage ILF3 high expression induced AAA progression
Bardoxolone methyl (BM, CDDO-Me) is an oral oleanolic acid-derived semi-synthetic triterpenoids and has been proven as an effective inducer of Keap1-Nrf2 (induce Keap1-Nrf2 disassociation, promotes Nrf2 protein stabilization and nuclear translocation 22 ) as well as an inhibitor of the NF-κB pathway(inhibit NF-κB p65 to the nucleus 23 ), which is undergoing clinical trials to explore the potential to treat chronic kidney diseases 24 , 25 . Given the shared targets and mechanisms between BM and ILF3, we aim to explore the potential of BM in treating AAA with high ILF3 expression, while simultaneously further validating the underlying mechanisms of ILF3’s action. Initially, we administered BM to ILF3 M-WT ApoE −/− mice and observed that BM treatment resulted in a reduction of AAA incidence, suggesting a protective effect of BM against the development of AAA (Supplementary Fig. 11 ). Furthermore, the in vitro research indicated that BM could restrain the inflammatory response induced by ILF3 overexpression via hindering NF-κB pathway and enhancing the Keap1-Nrf2 pathway (Supplementary Figs. 12A – 12F ). While no significant changes in blood pressure were observed across different treatment groups (Supplementary Table 4 ), BM potently attenuated exacerbated AAA progression in Ang II-treated ILF3 M-Tg ApoE −/− mice (Fig. 7A ). This was evidenced by reduced incidence of aneurysms (Fig. 7B ), rupture rate (Fig. 7C ), and mortality (Fig. 7D ) attributed to ectopic ILF3 expression. Vascular ultrasound imaging and maximal external aortic diameter measurement further corroborated BM’s inhibitory effects on vessel dilation in ILF3 M-Tg ApoE −/− mice (Figs. 7E, F ). Furthermore, BM treatment significantly ameliorates Ang II-induced elastin degradation (Figs. 7G, H ). The CaCl 2 -induced AAA model also revealed that BM treatment ameliorates ILF3 overexpression-caused abdominal aorta dilation (Figs. 7I, J ) and elastin disruption (Supplementary Figs. 12G and 12H ). In sum, the above results suggest that pharmacological intervention with BM may counteract the effects of macrophage ILF3 overexpression on NF-κB and Keap1-Nrf2 pathway and mitigate the detrimental effects of ILF3 overexpression in macrophages on AAA by restoring inflammatory homeostasis inflammatory homeostasis (Fig. 8 ).

A Schematic representation of study design and representative images showing the morphology of the whole aorta from all groups showing the macroscopic characteristics of aneurysms. B – D The AAA incidence ( B ) rupture rate ( C ) and Kaplan-Meir survival curve ( D ) across different groups (ILF3 M-WT ApoE −/− , n = 20; ILF3 M-Tg ApoE −/− and BM-treated ILF3 M-Tg ApoE −/− mice, n = 19). Statistical analyzes employed Fisher’s exact test with a two-tailed analysis for ( B , C ) and the Log-rank test for ( D ). E and F Representative ultrasound images ( E ) and aorta external diameter measurements ( F ) of Ang II-infused ILF3 M-WT ApoE −/− , ILF3 M-Tg ApoE −/− , and BM-treated ILF3 M-Tg ApoE −/− mice ( n = 9 per group). G and H Representative hematoxylin and eosin (HE) and Verhoeff-Van Gieson (VVG) staining ( G ) and elastin degradation grading ( H ) of the mouse aorta obtained from ILF3 M-WT ApoE −/− , ILF3 M-Tg ApoE −/− , and BM-treated ILF3 M-Tg ApoE −/− mice after AngII stimulus ( n = 10 per group; Scale bar=100 μm). I and J Schematic representation of study design and representative photographs ( I ) of saline-treated and CaCl 2 -induced aneurysm and maximal abdominal aortic diameter ( J ) n = 9 per group). Data are represented as mean ± SD. For comparisons between the three groups, one-way ANOVA followed by Tukey’s post hoc multiple comparisons test was used (7 F , 7 H , 7 J ). Boxplots were created using the first and third quartiles to define the bounds of the box, with the median shown in a circle. The minima are defined as the first quartile minus 1.5 times the inter-quartile range (IQR), and the maxima are defined as the third quartile plus 1.5 times the IQR. Violin plots display the density distribution of the data points through smoothed histograms.

Under the AAA pathological environment, ILF3 was upregulated in aneurysmal macrophages. Macrophage ILF3 activation accelerates the degradation of p105 mRNA and increases nuclear p65 translocation, intensifying pro-inflammatory responses via activation of the NF-κB pathway. On the other hand, macrophage ILF3 may bind to Keap1 mRNA via the protein complex ILF3-eIF4A1 and amplifies Keap1 translation, which contributes to the suppression of anti-inflammatory responses via restraining Keap1-Nrf2 activity. That destroys inflammatory homeostasis in focal AAA lesions, resulting in the development of AAA. Pharmacological therapy with bardoxolone methyl (BM) ameliorates ILF3 overexpression-induced AAA development via crosstalk with the NF-κB and Keap1-Nrf2 pathways.
The overarching findings of our study demonstrated that ILF3 is upregulated in aneurysmal macrophages. This upregulation of macrophage ILF3 appears to accelerate the formation of AAA, as corroborated by human and mouse AAA tissue data. Importantly, we found that the activation of macrophage ILF3 disrupts the inflammatory balance within localized AAA lesions. This disruption occurs through two primary mechanisms: enhancing the NF-κB pathway by destabilizing p105 mRNA and inhibiting the Keap1-Nrf2 pathway by increasing Keap1 translational efficiency via the ILF3/eIF4A1 complex. Moreover, our study highlights the therapeutic potential of BM in mitigating the adverse effects of AAA exacerbated by macrophage ILF3 activation.
Prior studies have shown that ILF3 is involved in the progression of cardiovascular diseases such as myocardial infarction 26 , 27 . Our previous work demonstrated that elevated ILF3 expression could promote arteriosclerotic calcification by acting on the promoter regions of BMP2 and STAT1 28 . A recent study revealed that ILF3 participates in the process of circACTA2 reducing CDK4 mRNA stability and protein expression via competing for association with CDK4 mRNA, thus leading to Ang II-induced VSMC death 15 . However, the specific role of ILF3 in AAA development is still unclear. Here, using bioinformatic and immunological methods, we have identified markedly elevated levels of ILF3 in AAA tissues of humans and mice. Notably, this increase was closely associated with a heightened accumulation of vascular macrophages, denoting the potential role of macrophage ILF3 in AAA progression.
The adventitial remodeling and inflammation are the main pathological features of AAA 9 , 29 . Macrophages are key phenotypes of immune cells in the adventitial, where they secrete pro-inflammatory mediators, including TNF-α, IL-6, and MCP-1 to initiate a positive feedback loop to extensive remodeling of the aortic wall and eventual aortic dissection 30 , 31 . Mellak et al. found that macrophage depletion inhibits the development of AAA using bone marrow transplantation in ApoE −/− mice 32 . However, the underlying molecular mechanisms that regulate macrophagic inflammatory homeostasis in situ are still to be comprehensively understood. Our study addresses this gap through a comprehensive approach involving bioinformatic and proteomic analyzes. We discovered a robust association between ILF3 and inflammatory responses, specifically implicating the NF-κB and Keap1-Nrf2 pathways. Our gene silencing experiments showed that inhibiting macrophage ILF3 led to a marked reduction in macrophage infiltration. The dampened inflammatory response was directly associated with a pronounced reduction in aortic remodeling and dissection in mouse models of AAA, thereby unequivocally establishing a causal link between ILF3 activity and AAA pathology. Furthermore, our in vitro and in vivo analyzes added another layer of understanding. We found that ILF3 deficiency mitigates the release of the chemokine MCP-1 and reduces the levels of pro-inflammatory cytokines such as IL-6 and TNF-α. Given that MCP-1 is a potent chemotactic factor for macrophage recruitment 33 and that IL-6 and TNF-α are critical players in vascular inflammation 34 , 35 , our findings have broad implications. It suggests that targeting ILF3 could disrupt multiple facets of the inflammatory cascade that contribute to AAA development, and further pharmacological targeting of ILF3 could disrupt multiple components of the inflammatory cascade, thereby offering a comprehensive approach to AAA treatment.
Compelling evidence underscores the pivotal role of NF-κB p65-mediated transactivation in the etiology of various chronic inflammatory diseases. Achieving targeted inhibition of p65, tailored to specific tissues and contexts, remains a significant therapeutic objective. In our research, the absence of macrophage ILF3 did not alter p65 levels, yet we observed an upregulation of p105 expression as confirmed by proteomic and immunoblotting analyzes. Previous studies have reported that p105 acts as an essential suppressor of inflammation, with p105 deficiency leading to spontaneous lymphocytic inflammation in the liver and lungs. Additionally, macrophages deficient in p50 have been shown to secrete increased pro-inflammatory cytokines 36 , 37 , 38 . However, the specific mechanisms by which p50/p105 regulates inflammatory responses in AAA lesions remain unclear. In our current study, we found that macrophage ILF3 deficiency caused an evident increase in cytoplasmic p105 and p50, as well as nuclear p50. Additionally, the results of Co-IP assays revealed elevated levels of p50:p105 and p50:p50 dimers, along with a decrease in p65:p50 dimers in ILF3-deficient macrophages, which accounted for the retention of cytoplasmic p65 and subsequent inhibition of inflammation. Furthermore, using RIP-seq and RT-PCR, we proved that ILF3 can bind to p105 mRNA, thus enhancing its degradation. As well as this, we also found that macrophage ILF3 enhances Keap1 protein expression, leading to decreased nuclear Nrf2. Mechanistically, RIP-seq, IP-MS and ribosome profile analysis unveiled that ILF3 augmented Keap1 translational efficiency by forming the ILF3/eIF4A1/Keap1 mRNA complex 39 . These results demonstrate the role of ILF3 in modulating inflammatory homeostasis and suggest that ILF3 can affect the translation efficiency of specific proteins via the formation of protein complexes.
The NF-κB and Keap1-Nrf2 pathways are vital in regulating inflammatory responses’ physiological homeostasis. Currently, there is an urgent need for therapeutics capable of targeting these two pathways. Herein, we revealed that ILF3 deficiency could inhibit NF-κB-DNA binding activity and increase Nrf2-DNA binding capacity, thus leading to a decreased inflammatory response. BM has been proven as an effective inducer of Keap1-Nrf2 and an inhibitor of the NF-κB pathway 40 , 41 . Growing evidence from the Bardoxolone Methyl Evaluation in Patients with Chronic Kidney Disease and Type 2 Diabetes (BEACON) trial and the Phase 2 Study of Bardoxolone Methyl in Patients with Chronic Kidney Disease and Type 2 Diabetes (TSUBAKI) study shows that BM not only can effectively increase eGFR and improve kidney function, but also can prevent cancer development and mitigate radiation-induced damage 24 , 42 , 43 . Our current investigation unveiled that BM counteracts the alterations in NF-κB and Keap1-Nrf2 DNA activity induced by ILF3 overexpression in vitro. This observation further substantiates the role of ILF3 in modulating the inflammatory response through its impact on the NF-κB and Keap1-Nrf2 signaling pathways. Notably, our in vivo assessments demonstrated that BM effectively attenuates the rapid progression of AAA associated with ectopic ILF3 expression, which offers a promising therapeutic direction for treating patients with AAA characterized by elevated ILF3 levels.
In conclusion, our study revealed the pivotal role of macrophage ILF3 on AAA development. We observed that in the pathological context of AAA, there is an upregulation of macrophage ILF3, which correlates with a worsening inflammatory response. Our mechanistic insights reveal that silencing macrophage ILF3 could offer arterial protection by mitigating inflammatory dilation. Specifically, macrophage ILF3 accelerates the degradation of p105 mRNA, enhancing the nuclear translocation of p65. This process activates the NF-κB pathway, leading to amplified pro-inflammatory responses. Moreover, ILF3’s interaction with Keap1, through the ILF3-eIF4A1 complex, augments Keap1 translation, which in turn dampens anti-inflammatory activities by restraining the Keap1-Nrf2 pathway. These mechanistic insights suggest that targeting macrophage ILF3, either through gene silencing or specific pharmacological treatments, presents a promising avenue for AAA therapy.
Microarray data collection and analysis
The NCBI-GEO database ( https://www.ncbi.nlm.nih.gov/geo/ ) was searched for microarray or RNA-sequencing data related to abdominal aortic aneurysm. We evaluated two studies, GSE57691 and GSE12591. Only normal aorta and abdominal aortic aneurysm samples were selected in each dataset. GSE57691 (Platform GPL570) included 10 control aortic specimen of organ donors or aortic specimen from 29 patients with AAA (mean maximum aortic diameter=68.4 ± 14.3 mm). GSE12591 (Platform GPL7215) comprised 3 whole aortas of 17 week old male ApoE − / − mice exposed to angiotensin II (1.44 µg/kg/min) for 4 weeks where there was clear evidence of aortic aneurysm formation and those exposed to saline infusion ( n = 3). We used the GEO2R online tool to analyze the raw data of microarrays and identify DEGs between the control tissues and AAA. GEO2R is an interactive web toll that allows users to compare different groups of samples in a GEO series to examine differentially expressed genes according to experimental conditions. Fold Change > 1.2 and P < 0.05 were used as the cut-off standards to obtain DEGs.
Single cell RNA-seq data processing
Sequencing data were acquired from the Gene Expression Omnibus (accession no. GSE166676). The Seurat package (Version 3.2.0) in R software (Version 4.2.1) was used for cross-sample adjustment, processing, and quality control. After removing low-quality cells, gene expression matrices were normalized using the Normalize Data function, and 2000 features with high cell-to-cell variation were identified using the Find Variable Features function. The Run PCA, Find Neighbors and Find Clusters functions were used to reduce dimensionality and cluster the cells, followed by dimensional reduction with the Run Uniform Manifold Approximation and Projection (UMAP) and Runtsne functions. By matching the cell cluster gene signature with the assumed cell type specific marker, the aggregated cells are mapped to the corresponding cell type.
The average gene expression level was calculated and the log-two-fold change between the specific cell cluster and other cells was applied as the test statistic. Enrichment analysis for the functions of differentially expressed genes (DEGs) was conducted using the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) functions in the Cluster Profiler in R; enrichment in GO terms and KEGG pathways were calculated based on hypergeometric distribution. The functions with p -value < 0.05 were considered significantly different.
Collection of human tissue samples
Human AAA tissues from ten male patients undergoing surgical repair for AAA in the Beijing Anzhen Hospital (N = 10) were donated by Dr. Wen-jian Jiang. The detailed information on the ten patients is listed in Extended Data Table S1 . The protocol was approved by local Ethics Committee of Beijing anzhen hospital capital medical university (#2018004). Patient consent was obtained for the use of this tissue. Age-matched non-AAA segments acquired from organ donors were harvested during explantation (N = 10) of Shandong provincial hospital. The protocol was endorsed by the Ethics Committee of Shandong First Medical University (#2022-303).
Animal models and pharmacological therapy
ILF3 Flox/Flox mice, which were generated by inserting the mouse ILF3-201 transcript between the first and second exons of the mouse ROSA26 locus. Specifically, the targeting vector used for this purpose was engineered to include a splice acceptor (SA), a loxP-flanked STOP cassette containing three polyadenylation signals, and the ILF3 cDNA, followed by an additional polyadenylation signal. This vector was introduced into C57BL/6 J zygotes via microinjection, along with Cas9 mRNA and sgRNA targeting the first intron of the Gt(ROSA)26Sor locus (ROSA26). The procedure was conducted by Viewsolid Biotech Co, Ltd, Beijing, China. These ILF3 Flox/Flox mice were hybridized with C57BL/6 LysMCre mice to generate macrophage-ILF3 conditional overexpression mice (ILF3 Flox/Flox LysMCre + ; hereafter referred to as ILF3 M-Tg ) and its littermates (ILF3 Flox/Flox LysMCre - ; hereafter referred to as ILF3 M-WT ). ILF3 flox/flox mice, possessing two loxP sites flanking exon 3 of the ILF3 gene (Viewsolid Biotech Co, Ltd; Beijing, China), were crossed with C57BL/6 LysMCre mice to generate macrophage-ILF3 knockout mice (ILF3 flox/flox LysMCre + ; hereafter referred to as ILF3 M-KO ) and controls (ILF3 flox/flox LysMCre - ). ApoE − / − mice were purchased from Beijing Viewsolid Biotechnology (Beijing, China) and crossed with ILF3 M-KO , ILF3 M-Tg , and their littermates to generate macrophage ILF3 genetic-engineering mice in ApoE − / − background. To avoid potential confounding factors related to sex-specific differences, male mice aged eight weeks old were selected in this study 44 , 45 . The mice were housed at 40 ± 5% humidity and 24 ± 2 °C under a 12 h light/dark cycle with access to water and food ad libitum.
To establish an angiotensin II (Ang II)-induced AAA mouse model, a mini osmotic pump (Alzet Model 2004, Alza Corp, Palo Alto, CA, USA) loaded with Ang II (1000 ng/kg/min, Sigma-Aldrich, Saint Louis, MO, USA) or saline (0.9% NaCl) was implanted subcutaneously at the dorsum of the neck for Ang-II or saline perfusion, and with concurrent high-fat feeding. Four weeks later, the abdominal aorta was collected for further analysis. Mouse aortic diameter was measured after cleaning the perivascular tissue using the microscope eyepiece reticle micrometer ruler, as mentioned in previous studies 46 .
To establish a CaCl 2 -induced AAA mouse model, infrarenal abdominal aortas were isolated and wrapped with fertile cotton balls presoaked in 0.5 mol/L calcium chloride for 15 min under general anesthesia using isoflurane. After four weeks, the mice were sacrificed for sample collection.
BM (CDDO-Me, Selleck, Shanghai, China) was dissolved in dimethyl sulfoxide (DMSO) and further diluted in PEG300 and PBS. Ang II-infused and CaCl 2 -treated mice were intraperitoneally injected with BM (1.25 mg/kg/day) or saline on the day after Ang II or CaCl 2 treatment 47 . Four weeks later, the mice were sacrificed for sample collection.
This study was conducted in strict compliance with the proposals outlined in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. All animal experiments have been reviewed and approved by the Institutional Animal Care and Use Committee of Shandong University (KYLL2020-454).
Blood pressure and weight measurement
Blood pressures were measured before and four weeks after Ang-II or saline perfusion using the pulse-based tail-cuff method with a photoelectric device (Softron, BP-2010). Mice were sacrificed four weeks after Ang-II or saline perfusion, and body weights were obtained.
Vascular ultrasound and quantification of AAA
Pulse wave velocity and aortic diameter were assessed using an ultrasound machine (Visualsonics, Vevo2100). Before positioning the collection probe, the mice anesthetized by inhalation of isoflurane were placed on the heated 37 °C plastic pillow, scratched with water ultrasonic transmission gel and coated on the mouse abdomen. Basal temperature and heart rate were observed throughout the ultrasound imaging procedure. Vascular ultrasonography was conducted to measure the maximal luminal diameter, with assessments independently carried out by two observers blinded to the experimental groups. Measurements were taken both the day before Ang II infusion and on day 28 post-infusion, as previously described 48 , 49 . B-mode imaging was used to determine the diameter of the suprarenal aorta and quantify the incidence of aneurysms according to the definition of AAA, which is defined by an enlargement of the aortic diameter by ≥50% compared to its baseline 50 , 51 . Furthermore, any dissection resulting in an intramural hematoma in the AAA mouse model should be considered, even if the dilation is just 110% 50 .
Macrophage isolation culture
Peritoneal macrophages were isolated through peritoneal lavage three days after the intraperitoneal injection of sterile suspension of 6% starch. Peritoneal macrophages were harvested, washed in phosphate-buffered saline (PBS), and cultured in DMEM medium with 1% penicillin/streptomycin and 10% bovine serum.
Bone marrow-derived macrophages were isolated from mouse femurs and tibiae, as described previously 52 . PBS was used to flush out the bone marrows of femurs and tibiae with a syringe with a 27 G needle. Cell suspension under 4 °C to 1500 rpm centrifuge for 5 min. Cells were then cultured with complete DMEM containing 30 ng/ml recombinant macrophage colony-stimulating factor (M-CSF) (R&D, Minnesota, USA) for macrophage differentiation. After 7 days, bone marrow-derived macrophages were acquired and used for further study.
Immunoblot analysis
Protein samples were electrophoresed on 10% sodium dodecyl sulfate-polyacrylamide gels and then transferred to a PVDF membrane. The membranes were blocked with 5% nonfat milk at room temperature for 1 h and then incubated with a specific diluted primary antibody overnight. After being washed 3 times with TBST, membranes were incubated with secondary antibodies at room temperature for 1 h. Chemiluminescence (Millipore, Billerica, MA, USA) identified the bound primary antibodies and measured using Image J (NIH, Bethesda, MD, USA). Antibodies used are as follows: β-actin, RRID: AB_2223172, Cell Signaling Technology, Catalog number: 4970; NF-κB1, RRID: AB_2282895, Cell Signaling Technology, Catalog number: 4717; NF-kappa B, RRID: AB_10859369, Cell Signaling Technology, Catalog number: 8242; NRF2, RRID: AB_2715528, Cell Signaling Technology, Catalog number: 12721; eIF4A1, RRID: AB_, Abcam, Catalog number: ab185946; IL-6, RRID: AB_2927381, Abcam, Catalog number: ab259341; MCP1, RRID: AB_448636, Abcam, Catalog number: ab25124; TNF-α, RRID: AB_2935774, Abcam, Catalog number: ab215188; Keap1, RRID: AB_, Abcam, Catalog number: ab227828; IL-10, RRID: AB_2847946, Abcam, Catalog number: ab189392; ILF3, RRID: AB_2049804, Abcam, Catalog number: ab92355; Lamin B1, RRID: AB_443298, Abcam, Catalog number: ab16048. Expression was normalized relative to the control.
Electrophoretic mobility shift assay (EMSA)
An EMSA was conducted using Light Shift Chemiluminescence EMSA kit (Thermo Fisher Scientific, Rockford, USA) to assess Ang II-induced NF-κB and Nrf2 activation in peritoneal macrophages. Nuclear extracts isolated from cultured peritoneal macrophages (5 × 10 6 /ml) using the NE-PER Nuclear and Cytoplasmic Extract Reagents (Thermo Fisher Scientific, Rockford, USA). The NF-κB and Nrf2 binding assay to predicted sites was conducted using annealed biotin labeled oligonucleotide probes. The labeled oligonucleotide probe designed for NF-κB is 5′(Biotin)-AGTTGAGGGGACTTTCCCAGGC-3′, and the probe for Nrf2 is 5′(Biotin)-TGGGGAACCTGTGCTGAGTCACTGGAG-3′.
RNA immunoprecipitation (RIP)
The RIP assay involved using the Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Billerica, MA, USA) in strict accordance with the manufacturer’s recommendations. Anti-ILF3 (19887-1-AP, Proteintech) and anti-IgG antibodies were used to immunoprecipitation RNAs. The final analysis was performed with qRT-PCR, showing fold enrichment of the target RNAs between the immunoprecipitated and input fractions.
Co-immunoprecipitation (Co-IP)
The cell lysates of macrophages were collected and centrifuged, followed by incubation with anti-P50 (SC-8414, Santa), anti-eIF4A1 (ab185946, Abcam), and anti-ILF3 (19887-1-AP, Proteintech) antibodies at 4 °C overnight. A lysate immunoprecipitated with anti-IgG (ab172730, Abcam) antibody was used as a negative control. The immunocomplexes were then purified using 30 µl of protein G magnetic beads (Bimake, Houston, USA) for 2 hours at 4 °C, centrifuged, and washed with Pierce TM IP Lysis Buffer (Thermo Fisher Scientific, Waltham, MA, USA) four to five times. The immunoprecipitated protein was further analyzed by western blot with target antibodies.
Histological analysis
Abdominal aortas were harvested, perfused with saline, and then fixed with 4% paraformaldehyde, followed by embedding in paraffin and serial sectioning. Paraffin-embedded sections (5 µm thick) were prepared for hematoxylin and eosin (HE) staining to observe general morphology and Verhoff-Van Gieson (VVG) staining to evaluate elastin degradation. Elastin degradation was graded using a quantification method by counting the number of breaks per vessel 3 .
Immunohistochemical and immunofluorescence staining
The paraffin embedded arterial sections were dewaxed and stained with primary antibodies, followed by a biotin-conjugated secondary antibody (1:500) and horseradish peroxidase-conjugated streptavidin (Dianova, Rodeo, CA).
For immunofluorescence staining, sections were stained overnight with specific antibodies at 4 °C and then added with fluorescence-labeled secondary antibodies. Nuclei were stained with 4,6-diamidino-2-phenylindole (DAPI). Specific fluorescence imaging was obtained by laser-scanning confocal microscopy (LSM710, Carl Zeiss, Germany).
Immunoprecipitation mass spectrometry (IP-MS)
IP-MS was conducted as previously described 53 . Briefly, peritoneal macrophages were obtained from C57BL/6 mice and separated. IP was performed using an immunoprecipitation kit (Abcam, Cambridge, UK). In brief, cells were collected, resuspended, and lysed in lysis buffer containing the protease inhibitor cocktail. The resulting cell lysates and anti-ILF3 antibody (19887-1-AP, Proteintech) were mixed overnight in a rotary mixer at 4 °C. Next, 40 µL protein A sepharose bead slurry was added into each tube and incubated for 1 h at 4 °C, followed by 2000 g centrifugation for 2 min. After washing three times with 1 mL 1× wash buffer, 40 µL 2× SDS-PAGE loading buffer added to the beads and boiled for 5 min to elute the complex.
Western blot analysis was used for the validation of IP samples. The gels were then excised and subjected to MS. UPLC-MS/MS involved using nano ACQUITY UPLC Columns (Waters) with Orbitrap Elite Mass Spectrometers (Thermo Fisher Scientific, Waltham, MA, USA). The resulting MS/MS data were handled with Proteome Discoverer 1.3 (high peptide confidence, peptide ion score > 20).
Flow cytometry
To identify subsets of inflammatory cells in murine models, single-cell suspensions were isolated from mouse aortas following established protocols 54 . The enzymatically digested suspensions were filtered through a 70 µm sterile mesh and centrifuged at 300 g for 5 minutes at 4 °C. The resulting pellet was resuspended in a buffer containing 0.5% BSA. Cell viability and count were determined using trypan blue exclusion and an automated cell counter (Countess, Life Technologies). Anti-CD16/CD32 (101302, biolgend) was added and incubated for 10 minutes at room temperature to minimize nonspecific binding. Subsequently, cells were stained with specific marker antibodies APC anti-mouse CD83(121509, biolgend), Percpcy5.5 anti-mouse CD45(103132, biolgend), FITC anti-mouse CD11b(101206, biolgend), PE anti-mouse F4/80(12-4801-82, ebioscience), Pacific Blue anti-mouse Ly-6G/Ly-6C(Gr-1)(108429, biolgend), PE anti-mouse CD19(115507, biolgend), APC anti-mouse NK1.1(108709, biolgend) and FITC anti-mouse CD3 (100203, biolgend). Data were further analyzed by NovoExpress® Software.
The enzyme-linked immunosorbent assay (Elisa)
After modeling, blood samples were collected and stored at −80 °C until further analysis. Following the manufacturers’ guidelines, plasma levels of IL-6 (Cat#88-7064, Invitrogen), IL-10 (Cat#88-7105, Invitrogen), TNF-α (Cat#88-7324, Invitrogen), and MCP-1 (Cat#BMS6005, Invitrogen) were quantified using Elisa kits.
Total cellular protein was extracted from isolated peritoneal macrophages derived from both ILF3 M-KO mice ( n = 3) and wild-type counterparts ( n = 3) for analysis through 4D label-free quantification (4D-LFQ) proteomics, conducted by PTM Biolabs (Hangzhou, China). The analysis employed a false discovery rate (FDR) of 5% for both protein identification and peptide spectral matching (PSM), establishing a threshold for differential expression: proteins with fold changes exceeding 1.2 and adjusted P values below 0.05 were classified as differentially expressed proteins (DEPs). Functional enrichment analysis of these DEPs was carried out using a two-tailed Fisher’s exact test. Significance was attributed to Gene Ontology (GO) terms, KEGG pathways, and protein domains that achieved a p -value of less than 0.05.
Ribosome sequencing (Ribo-seq) and RNA Sequencing
Ribosome profiling was performed on isolated peritoneal macrophages from both ILF3 M-KO ( n = 6) and ILF3 M-WT ( n = 6) tissues by Gene Denovo (Guangzhou, China). In brief, cycloheximide (Sigma, St Louis, MO, USA; 100 µg/ml) was added to the peritoneal macrophage medium for 2 min to block translational elongation. The supernatant was collected after trituration and centrifuged at 20000 g for 10 min at 4 °C. To prepare RFs, 10 µL of RNase I (NEB, Ipswich, MA, USA) and 6 µL of DNase I (NEB, Ipswich, MA, USA) were added to 400 µL of lysate, mixed and incubated for 45 min at room temperature. Nuclease digestion was halted via adding 10 µL of SUPERase, an RNase Inhibitor (Ambion, Austin, TX, USA). Then 100 μL of digested RFs was added to the prepared Size exclusion columns (Illustra MicroSpin S-400 HR Columns; GE Healthcare; catalog no. 27- 5140-01) and centrifuged (600 g, 2 min). Subsequently, 10 μL of 10% (w/v) SDS was added up to the elution, and RFs (size > 17nt) were separated via RNA Clean and Concentrator-25 kit (Zymo Research; R1017). Further, rRNA and residual DNA probes were removed via RNase H (NEB, Ipswich, MA, USA) and DNase I (NEB, Ipswich, MA, USA). Lastly, the RFs were refined by magnet beads (Vazyme, Nanjing, Jiangsu, China).
Ribo-seq libraries were framed by the NEBNext® Multiple Small RNA Library Prep Set for Illumina® (catalog no. E7300S, E7300L). In brief, adapters were added up to RFs, succeeded by reverse transcription and PCR amplification. The PCR products (140-160 bp size) were enriched to construct cDNA library, sequenced using Illumina HiSeq TM X10. Moreover, we used FASTP to filter the raw data and remove low-quality bases (over 50%) or N bases (over 10%). After trimming the adapter sequences, reads with length between 20–40 bp were kept for analysis. Raw reads were filtered and mapped to the mouse ribosome RNA database using bowtie2. The reads mapped to rRNAs, transfer RNAs (tRNA), small nuclear RNAs (snRNAs), small nucleolar RNAs (snoRNAs), and miRNAs were eliminated. Processed RNA-seq reads were mapped to the rn6 genome using whole-genome alignment by STAR with a 2-pass setting. Ribo-seq data analysis was performed as previously described.[23] Differential translated genes between groups were defined as |log2(FoldChange)| > 0.3 and p -value < 0.05.
For RNA-seq, total RNA of peritoneal macrophages from both ILF3 M-KO ( n = 6) and ILF3 M-WT ( n = 6) tissues was isolated utilizing the RNeasy Mini Kit (Qiagen, Germany). Subsequently, strand-specific libraries were constructed employing the TruSeq® Stranded Total RNA Sample Preparation kit (Illumina, USA) following the manufacturer’s instructions. Library quantification was conducted using a Qubit® 2.0 Fluorometer (Life Technologies, USA) and validated by Agilent 2100 bioanalyzer (Agilent Technologies, USA) to confirm the insert size and calculate the mole concentration. Library clustering was performed on a cBot system at a concentration of 10 pM, followed by sequencing on the HiSeq X Ten platform (Illumina, USA) (Illumina, USA). The sequencing raw reads were preprocessed to remove rRNA sequences, sequencing adapters, short fragments, and other low-quality reads. We employed Tophat v2.0.9 55 for mapping the filtered reads to the mouse mm10 reference genome, allowing for a maximum of two mismatches. Following genomic mapping, Cufflinks v2.1.1 56 was utilized with a reference annotation to calculate FPKM values for known gene models. Differential expression of genes between the groups was ascertained using the DESeq R package, with stringent criteria set at FoldChange > 2 and p -value < 0.05. Translational efficiency was quantified by the ratio of translating mRNAs to total mRNAs, defined as: TE = (FPKM in Ribo-seq) / (FPKM in RNA-seq). Furthermore, to elucidate the functional similarity among identified genes, we employed the Friends analysis, a method executed using the GOSemSim package 57 . The raw data from the ribosome sequencing and associated mRNA sequencing conducted in this study are publicly available, which have been deposited in the NCBI Gene Expression Omnibus (GEO) and can be accessed through the GEO Series accession numbers GSE241911, GSE242024, GSE242088, and GSE242185.
mRNA stability analysis and qRT-PCR
Macrophages were suspended in TRIzol Reagent (Invitrogen, Waltham, MA, USA) after treatment with actinomycin D (Act-D, 10 mg/mL) for 0, 2, 4, 8, and 12 h. Total RNA was purified using the RNeasy Mini Kit (Qiagen, Hilden, Germany) based on the manufacturer’s recommendations. The total RNA was reverse transcribed by using the PrimeScript RT Reagent Kit with gDNA Eraser (Perfect Real Time) (Takara, Japan), and qRT-PCR was conducted using the LightCycler 480 SYBR Green I Master (Roche Life Science, NSW, Australia). The level of each transcript was normalized to the control gene β-actin and shown relative to the average expression of the control sample ( n = 5 under each experimental condition).
Statistical analysis
The ggplot2 package of the interesting plots in the finding of the R foundation and statistic in R v4.2.1 (R Foundation for Statistical Computing, Vienna, Austria) 58 , 59 , and Prism 9.0 (GraphPad Inc) were used for statistical analyzes. Continuous parameters are presented as (mean ± SD), and categorical variables are shown as percentages. After verification of the normal distribution of the data via the Shapiro-Wilk test and variance equality via the Levene test, the Student’s t test was used when the variance of the two groups was equal, and the unequal variance t test was used for data with unequal variances. Otherwise, the Mann-Whitney U test is for data sets that are not normally distributed. Brown Forsythe test was used to evaluate the homogeneity of variance in multiple groups ( > 2 groups). After One-way ANOVA analysis, a Tukey post-hoc analysis was used if the data passed the equal variance test; otherwise, a Welch ANOVA test was performed, followed by a post hoc analysis. Two-way ANOVA with a Bonferroni post hoc test was used to test the difference among groups with multiple factors. The survival curve analysis was analyzed using the Kaplan–Meier product-limit approach and compared by the log-rank test. We downloaded publicly available data from the Gene Expression Synthesis (GEO) database (GSE57691 and GSE12591) and obtained DEGs by GEO2R analysis.
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the iProX partner repository with the dataset identifier PXD047695. IP-MS datasets are available via ProteomeXchange with identifiers PXD047694. All sequencing data generated in this study have been deposited in the NCBI Sequence Read Archive under the BioProject ID PRJNA1050581 for Ribo-seq, PRJNA1050631 for RIP-seq. All other data are available in the article and its Supplementary files or from the corresponding author upon request. Source data are provided with this paper.
Sakalihasan, N. et al. Abdominal aortic aneurysms. Nat. Rev. Dis. Prim. 4 , 34 (2018).
Article PubMed Google Scholar
Shimizu, K., Mitchell, R. N. & Libby, P. Inflammation and cellular immune responses in abdominal aortic aneurysms. Arterioscler Thromb. Vasc. Biol. 26 , 987–994 (2006).
Article CAS PubMed Google Scholar
Liu, C. L. et al. Adipocytes promote interleukin-18 binding to its receptors during abdominal aortic aneurysm formation in mice. Eur. Heart J. 41 , 2456–2468 (2020).
Filiberto, A. C. et al. Endothelial pannexin-1 channels modulate macrophage and smooth muscle cell activation in abdominal aortic aneurysm formation. Nat. Commun. 13 , 1521 (2022).
Article ADS CAS PubMed PubMed Central Google Scholar
Wang, S. K. & Murphy, M. P. Immune modulation as a treatment for abdominal aortic aneurysms. Circ. Res 122 , 925–927 (2018).
Article CAS PubMed PubMed Central Google Scholar
Wallinder, J., Bergqvist, D. & Henriksson, A. E. Proinflammatory and anti-inflammatory cytokine balance in patients with abdominal aortic aneurysm and the impact of aneurysm size. Vasc. Endovasc. Surg. 43 , 258–261 (2009).
Article Google Scholar
Vucevic, D. et al. Inverse production of IL-6 and IL-10 by abdominal aortic aneurysm explant tissues in culture. Cardiovasc Pathol. 21 , 482–489 (2012).
Gavrila, D. et al. Vitamin E inhibits abdominal aortic aneurysm formation in angiotensin II-infused apolipoprotein E-deficient mice. Arterioscler Thromb. Vasc. Biol. 25 , 1671–1677 (2005).
Maiellaro, K. & Taylor, W. R. The role of the adventitia in vascular inflammation. Cardiovasc Res . 75 , 640–648 (2007).
Castella, S., Bernard, R., Corno, M., Fradin, A. & Larcher, J. C. Ilf3 and NF90 functions in RNA biology. Wiley Interdiscip. Rev. RNA 6 , 243–256 (2015).
Nazitto, R. et al. ILF3 is a negative transcriptional regulator of innate immune responses and myeloid dendritic cell maturation. J. Immunol. 206 , 2949–2965 (2021).
Li, K. et al. ILF3 is a substrate of SPOP for regulating serine biosynthesis in colorectal cancer. Cell Res . 30 , 163–178 (2020).
Yamada, Y., Matsui, K., Takeuchi, I. & Fujimaki, T. Association of genetic variants with dyslipidemia and chronic kidney disease in a longitudinal population-based genetic epidemiological study. Int J. Mol. Med . 35 , 1290–1300 (2015).
Yoshida, T. et al. Association of polymorphisms of BTN2A1 and ILF3 with myocardial infarction in Japanese individuals with or without hypertension, diabetes mellitus or chronic kidney disease. Int J. Mol. Med 27 , 745–752 (2011).
CAS PubMed Google Scholar
Ma, Y. et al. circACTA2 mediates Ang II-induced VSMC senescence by modulation of the interaction of ILF3 with CDK4 mRNA. Aging (Albany NY) 13 , 11610–11628 (2021).
Davis, F. M. et al. Inhibition of macrophage histone demethylase JMJD3 protects against abdominal aortic aneurysms. J. Exp. Med. 218 , e20201839 (2021).
Raffort, J. et al. Monocytes and macrophages in abdominal aortic aneurysm. Nat. Rev. Cardiol. 14 , 457–471 (2017).
Dorrington, M. G. & Fraser, I. D. C. NF-κB Signaling in Macrophages: Dynamics, Crosstalk, and Signal Integration. Front Immunol. 10 , 705 (2019).
Ahmed, S. M., Luo, L., Namani, A., Wang, X. J. & Tang, X. Nrf2 signaling pathway: Pivotal roles in inflammation. Biochim Biophys. Acta Mol. Basis Dis. 1863 , 585–597 (2017).
Liu, T., Zhang, L., Joo, D. & Sun, S. C. NF-κB signaling in inflammation. Signal Transduct. Target Ther. 2 , 17023 (2017).
Article PubMed PubMed Central Google Scholar
Armaos, A. et al. catRAPID omics v2.0: going deeper and wider in the prediction of protein-RNA interactions. Nucleic Acids Res. 49 , W72–w79 (2021).
Yang, J. L. et al. Keap1-Nrf2 signaling activation by bardoxolone-methyl ameliorates high glucose-induced oxidative injury in human umbilical vein endothelial cells. Aging (Albany NY) 12 , 10370–10380 (2020).
Wang, Y. Y., Yang, Y. X., Zhe, H., He, Z. X. & Zhou, S. F. Bardoxolone methyl (CDDO-Me) as a therapeutic agent: an update on its pharmacokinetic and pharmacodynamic properties. Drug Des. Devel Ther. 8 , 2075–2088 (2014).
PubMed PubMed Central Google Scholar
de Zeeuw, D. et al. Bardoxolone methyl in type 2 diabetes and stage 4 chronic kidney disease. N. Engl. J. Med . 369 , 2492–2503 (2013).
Shelton, L. M., Park, B. K. & Copple, I. M. Role of Nrf2 in protection against acute kidney injury. Kidney Int 84 , 1090–1095 (2013).
Yamada, Y. et al. Association of a polymorphism of BTN2A1 with myocardial infarction in East Asian populations. Atherosclerosis 215 , 145–152 (2011).
Yoshida, T. et al. Association of polymorphisms of BTN2A1 and ILF3 with myocardial infarction in Japanese individuals with different lipid profiles. Mol. Med. Rep. 4 , 511–518 (2011).
Xie, F. et al. ILF3 is responsible for hyperlipidemia-induced arteriosclerotic calcification by mediating BMP2 and STAT1 transcription. J. Mol. Cell Cardiol. 161 , 39–52 (2021).
Li, H. et al. Lysyl hydroxylase 1 (LH1) deficiency promotes angiotensin II (Ang II)-induced dissecting abdominal aortic aneurysm. Theranostics 11 , 9587–9604 (2021).
Sakaue, T. et al. Perivascular adipose tissue angiotensin ii type 1 receptor promotes vascular inflammation and aneurysm formation. Hypertension 70 , 780–789 (2017).
Wu, M. Y., Li, C. J., Hou, M. F. & Chu, P. Y. New insights into the role of inflammation in the pathogenesis of atherosclerosis. Int J. Mol. Sci. 18 , 2034 (2017).
Mellak, S. et al. Angiotensin II mobilizes spleen monocytes to promote the development of abdominal aortic aneurysm in Apoe-/- mice. Arterioscler Thromb. Vasc. Biol. 35 , 378–388 (2015).
Harada, T. et al. Focal adhesion kinase promotes the progression of aortic aneurysm by modulating macrophage behavior. Arterioscler Thromb. Vasc. Biol. 37 , 156–165 (2017).
Hiromi, T. et al. Excessive EP4 signaling in smooth muscle cells induces abdominal aortic aneurysm by amplifying inflammation. Arterioscler Thromb. Vasc. Biol. 40 , 1559–1573 (2020).
Dale, M. A., Ruhlman, M. K. & Baxter, B. T. Inflammatory cell phenotypes in AAAs: their role and potential as targets for therapy. Arterioscler Thromb. Vasc. Biol. 35 , 1746–1755 (2015).
Yu, Y., Wan, Y. & Huang, C. The biological functions of NF-kappaB1 (p50) and its potential as an anti-cancer target. Curr. Cancer Drug Targets 9 , 566–571 (2009).
Saccani, A. et al. p50 nuclear factor-kappaB overexpression in tumor-associated macrophages inhibits M1 inflammatory responses and antitumor resistance. Cancer Res. 66 , 11432–11440 (2006).
Cao, S., Zhang, X., Edwards, J. P. & Mosser, D. M. NF-kappaB1 (p50) homodimers differentially regulate pro- and anti-inflammatory cytokines in macrophages. J. Biol. Chem. 281 , 26041–26050 (2006).
Li, Z. et al. USP9X controls translation efficiency via deubiquitination of eukaryotic translation initiation factor 4A1. Nucleic Acids Res 46 , 823–839 (2018).
Chien, J. Y., Chou, Y. Y., Ciou, J. W., Liu, F. Y. & Huang, S. P. The effects of two nrf2 activators, bardoxolone methyl and omaveloxolone, on retinal ganglion cell survival during ischemic optic neuropathy. Antioxid. (Basel) 10 , 1466 (2021).
Article CAS Google Scholar
Yang, C. C., Lin, C. C., Jou, M. J., Hsiao, L. D. & Yang, C. M. RTA 408 inhibits interleukin-1β-induced MMP-9 expression via suppressing protein kinase-dependent NF-κB and AP-1 activation in rat brain astrocytes. Int J. Mol. Sci. 20 , 2826 (2019).
Sun, Q. et al. Bardoxolone and bardoxolone methyl, two Nrf2 activators in clinical trials, inhibit SARS-CoV-2 replication and its 3C-like protease. Signal Transduct. Target Ther. 6 , 212 (2021).
Nangaku, M. et al. Randomized clinical trial on the effect of bardoxolone methyl on GFR in diabetic kidney disease patients (TSUBAKI study). Kidney Int Rep. 5 , 879–890 (2020).
Golledge, J., Thanigaimani, S., Powell, J. T. & Tsao, P. S. Pathogenesis and management of abdominal aortic aneurysm. Eur. Heart J. 44 , 2682–2697 (2023).
Ailawadi, G. et al. Gender differences in experimental aortic aneurysm formation. Arterioscler Thromb. Vasc. Biol. 24 , 2116–2122 (2004).
Ding, Y. N. et al. SIRT6 is an epigenetic repressor of thoracic aortic aneurysms via inhibiting inflammation and senescence. Signal Transduct. Target Ther. 8 , 255 (2023).
Article CAS PubMed Central Google Scholar
Hisamichi, M. et al. Role of bardoxolone methyl, a nuclear factor erythroid 2-related factor 2 activator, in aldosterone- and salt-induced renal injury. Hypertens. Res. 41 , 8–17 (2018).
Barisione, C. et al. Rapid dilation of the abdominal aorta during infusion of angiotensin II detected by noninvasive high-frequency ultrasonography. J. Vasc. Surg. 44 , 372–376 (2006).
Shridas, P. et al. Adipocyte-derived serum amyloid a promotes angiotensin II-induced abdominal aortic aneurysms in obese C57BL/6J mice. Arterioscler Thromb. Vasc. Biol. 42 , 632–643 (2022).
Daugherty, A. & Cassis, L. A. Mouse models of abdominal aortic aneurysms. Arterioscler Thromb. Vasc. Biol. 24 , 429–434 (2004).
Trachet, B., Fraga-Silva, R. A., Jacquet, P. A., Stergiopulos, N. & Segers, P. Incidence, severity, mortality, and confounding factors for dissecting AAA detection in angiotensin II-infused mice: a meta-analysis. Cardiovasc Res. 108 , 159–170 (2015).
Toda, G., Yamauchi, T., Kadowaki, T. & Ueki, K. Preparation and culture of bone marrow-derived macrophages from mice for functional analysis. STAR Protoc. 2 , 100246 (2021).
ten Have, S., Boulon, S., Ahmad, Y. & Lamond, A. I. Mass spectrometry-based immuno-precipitation proteomics - the user’s guide. Proteomics 11 , 1153–1159 (2011).
Da Ros, F. et al. Targeting interleukin-1β protects from aortic aneurysms induced by disrupted transforming growth factor β signaling. Immunity 47 , 959–973 (2017).
Jackson, R. et al. The translation of non-canonical open reading frames controls mucosal immunity. Nature 564 , 434–438 (2018).
Trapnell, C. et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28 , 511–515 (2010).
Yu, G. et al. GOSemSim: an R package for measuring semantic similarity among GO terms and gene products. Bioinformatics 26 , 976–978 (2010).
Allen, M., Poggiali, D., Whitaker, K., Marshall, T. R. & Kievit, R. A. Raincloud plots: a multi-platform tool for robust data visualization. Wellcome Open Res. 4 , 63 (2019).
Wickham, H. ggplot2. WIREs Computational Stat. 3 , 180–185 (2011).
Download references
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 82370455 and 82200466), the National Key R & D Program of China (No.2017YFC0908700, 2017YFC0908703 and 2017YFC1308000), the Taishan Scholar Project of Shandong Province of China (No.ts20190972, No.tsqn202306377), Natural Science Foundation of Shandong Province (No.ZR2022QH296), Key Clinical Research project of Clinical Research Center of Shandong University (No.2020SDUCRCA016), and Academic promotion program of Shandong First Medical University (No.2021QL021).
Author information
These authors contributed equally: Zhao-yang Wang, Jie Cheng, Ying Wang, Hai-tao Yuan, Shao-jie Bi.
Authors and Affiliations
The Key Laboratory of Cardiovascular Remodeling and Function Research, Chinese Ministry of Education, Chinese National Health Commission and Chinese Academy of Medical Sciences, Department of Cardiology, Qilu Hospital of Shandong University, Jinan, China
Zhao-yang Wang, Jie Cheng, Ying Wang, Shuang-xi Wang, Ya-min Hou, Xu Zhang, Bo-han Xu, Ze-ying Wang, Yun Zhang & Ming-xiang Zhang
Department of Cardiology, Shandong Provincial Hospital Affiliated to Shandong First Medical University, Jinan, Shandong, China
Zhao-yang Wang & Hai-tao Yuan
Department of Cardiology, the Second Hospital, Cheeloo College of Medicine, Shandong University, Jinan, Shandong, China
Shao-jie Bi
Department of Cardiovascular Surgery, Beijing Anzhen Hospital, Capital Medical University, Beijing, China
Wen-jian Jiang
Department of Emergency and Chest Pain Center, Qilu Hospital, Shandong University, Jinan, China
Yu-guo Chen
You can also search for this author in PubMed Google Scholar
Contributions
Z.W., J.C., Y.W., H.Y., and S.B. contributed equally to this work. Z.W., M.Z., and Y.C. conceived the project. Z.W. and M.Z. wrote the manuscript. Y.Z., W.J., and M.Z. supervised the study. Z.W., J.C., H.Y., S.B., S.W., and Ze.W. performed human studies and carried out analyses. Z.W., J.C., Y.W., B.X., X.Z., and Y.H. performed animal experiments and prepared figures. Z.W., J.C., and M.Z. interpreted the results. All authors reviewed the results and approved the final version of the manuscript.
Corresponding authors
Correspondence to Yun Zhang , Wen-jian Jiang , Yu-guo Chen or Ming-xiang Zhang .
Ethics declarations
Competing interests.
The authors declare no competing interests.
Peer review
Peer review information.
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information, peer review file, description of additional supplementary files, supplementary data 1, supplementary data 2, reporting-summary, source data, rights and permissions.
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/ .
Reprints and permissions
About this article
Cite this article.
Wang, Zy., Cheng, J., Wang, Y. et al. Macrophage ILF3 promotes abdominal aortic aneurysm by inducing inflammatory imbalance in male mice. Nat Commun 15 , 7249 (2024). https://doi.org/10.1038/s41467-024-51030-4
Download citation
Received : 03 December 2023
Accepted : 24 July 2024
Published : 23 August 2024
DOI : https://doi.org/10.1038/s41467-024-51030-4
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
By submitting a comment you agree to abide by our Terms and Community Guidelines . If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Sign up for the Nature Briefing newsletter — what matters in science, free to your inbox daily.

A randomized double-blind placebo-controlled clinical trial of Guanfacine Extended Release for aggression and self-injurious behavior associated with Prader-Willi Syndrome
- Find this author on Google Scholar
- Find this author on PubMed
- Search for this author on this site
- ORCID record for Theresa Jacob
- For correspondence: [email protected]
- Info/History
- Preview PDF
Introduction: Prader-Willi Syndrome (PWS), a rare genetic disorder, affects development and behavior, frequently resulting in self-injury, aggression, hyperphagia, oppositional behavior, impulsivity and over-activity causing significant morbidity. Currently, limited therapeutic options are available to manage these neuropsychiatric manifestations. The aim of this clinical trial was to assess the efficacy of guanfacine-extended release (GXR) in reducing aggression and self-injury in individuals with PWS. Trial Design: Randomized, double-blind, placebo-controlled trial conducted under IRB approval. Methods: Subjects with a diagnosis of PWS, 6-35 years of age, with moderate to severe aggressive and/or self-injurious behavior as determined by the Clinical Global Impression (CGI)-Severity scale, were included in an 8-week double-blind, placebo-controlled, fixed-flexible dose clinical trial of GXR, that was followed by an 8-week open-label extension phase. Validated behavioral instruments and physician assessments measured the efficacy of GXR treatment, its safety and tolerability. Results: GXR was effective in reducing aggression/agitation and hyperactivity/noncompliance as measured by the Aberrant Behavior Checklist (ABC) scales (p=0.03). Overall aberrant behavior scores significantly reduced in the GXR arm. Aggression as measured by the Modified Overt Aggression Scale (MOAS) also showed a significant reduction. Skin-picking lesions as measured by the Self Injury Trauma (SIT) scale decreased in response to GXR. No serious adverse events were experienced by any of the study participants. Fatigue /sedation was the only adverse event significantly associated with GXR. The GXR group demonstrated significant overall clinical improvement as measured by the CGI-Improvement (CGI-I) scale. (p<0.01). Conclusion: Findings of this pragmatic trial strongly support the use of GXR for treatment of aggression, skin picking, and hyperactivity in children, adolescents, and adults with PWS. Trial Registration: ClinicalTrials.gov Identifier - NCT05657860
Competing Interest Statement
I have read the journal's policy and the authors of this manuscript have the following competing interests: DS has served as a consultant to Soleno Therapeutics, Acadia Pharmaceuticals, Tonix Pharmaceuticals, and Consynance Therapeutics. MS and TJ have no other competing interests to report.
Clinical Trial
ClinicalTrials.gov identifier: NCT05657860
Funding Statement
Author declarations.
I confirm all relevant ethical guidelines have been followed, and any necessary IRB and/or ethics committee approvals have been obtained.
The details of the IRB/oversight body that provided approval or exemption for the research described are given below:
This study was approved by the Institutional Review Board of Maimonides Medical Center (# 2020-11-03-MMC). Written, IRB-approved informed consent was obtained from each participant's parent or legal guardian, and assent was obtained from each participant, as applicable.
I confirm that all necessary patient/participant consent has been obtained and the appropriate institutional forms have been archived, and that any patient/participant/sample identifiers included were not known to anyone (e.g., hospital staff, patients or participants themselves) outside the research group so cannot be used to identify individuals.
I understand that all clinical trials and any other prospective interventional studies must be registered with an ICMJE-approved registry, such as ClinicalTrials.gov. I confirm that any such study reported in the manuscript has been registered and the trial registration ID is provided (note: if posting a prospective study registered retrospectively, please provide a statement in the trial ID field explaining why the study was not registered in advance).
I have followed all appropriate research reporting guidelines, such as any relevant EQUATOR Network research reporting checklist(s) and other pertinent material, if applicable.
Data Availability
All relevant data are within the manuscript and its Supporting Information files and will be available upon its publication.
View the discussion thread.
Thank you for your interest in spreading the word about medRxiv.
NOTE: Your email address is requested solely to identify you as the sender of this article.

Citation Manager Formats
- EndNote (tagged)
- EndNote 8 (xml)
- RefWorks Tagged
- Ref Manager
- Tweet Widget
- Facebook Like
- Google Plus One
Subject Area
- Psychiatry and Clinical Psychology
- Addiction Medicine (342)
- Allergy and Immunology (665)
- Anesthesia (180)
- Cardiovascular Medicine (2625)
- Dentistry and Oral Medicine (314)
- Dermatology (222)
- Emergency Medicine (397)
- Endocrinology (including Diabetes Mellitus and Metabolic Disease) (930)
- Epidemiology (12175)
- Forensic Medicine (10)
- Gastroenterology (756)
- Genetic and Genomic Medicine (4064)
- Geriatric Medicine (385)
- Health Economics (676)
- Health Informatics (2625)
- Health Policy (997)
- Health Systems and Quality Improvement (979)
- Hematology (360)
- HIV/AIDS (845)
- Infectious Diseases (except HIV/AIDS) (13659)
- Intensive Care and Critical Care Medicine (790)
- Medical Education (398)
- Medical Ethics (109)
- Nephrology (430)
- Neurology (3832)
- Nursing (209)
- Nutrition (570)
- Obstetrics and Gynecology (734)
- Occupational and Environmental Health (690)
- Oncology (2008)
- Ophthalmology (581)
- Orthopedics (238)
- Otolaryngology (304)
- Pain Medicine (250)
- Palliative Medicine (73)
- Pathology (471)
- Pediatrics (1107)
- Pharmacology and Therapeutics (459)
- Primary Care Research (447)
- Psychiatry and Clinical Psychology (3400)
- Public and Global Health (6499)
- Radiology and Imaging (1390)
- Rehabilitation Medicine and Physical Therapy (806)
- Respiratory Medicine (869)
- Rheumatology (400)
- Sexual and Reproductive Health (407)
- Sports Medicine (338)
- Surgery (441)
- Toxicology (52)
- Transplantation (185)
- Urology (165)

Study findings may help explain why we keep getting colds
Media Contact: Leila Gray - [email protected], 206-475-9809

The most common cause of the common cold, the rhinovirus, increases its chances of infecting someone who lacks immunity by simultaneously circulating many versions of itself, according to new research from the University of Washington School of Medicine in Seattle.
“With viruses like SARS-CoV-2 or influenza, one variant will dominate for a while and then another takes over, and it, in turn, is replaced by another,” said Dr. Alex Greninger , professor of laboratory medicine and pathology, who led the study. “Rhinovirus, on the other hand, appears to flood the zone with many discrete variants circulating in the community at the same time. It’s a way to overcome your defenses with sheer numbers.”
The findings were published in The Journal of Infectious Diseases.
Although the rhinovirus is the most common cause of upper respiratory infections, relatively little is known about how it evolves and spreads. In their study, the researchers analyzed nasal swabs taken at Seattle-area collection sites during the COVID-19 pandemic.

They looked at samples gathered during two periods:
- spring and summer of 2021, when strict COVID-19 public health measures slowed the spread of SARS-CoV-2 and nearly eliminated other common respiratory infections such as influenza and respiratory syncytial virus (RSV).
- fall and winter of 2022, when restrictions were relaxed, and influenza and RSV reemerged.
In 2021, while measures slowed the spread of SARS-CoV-2, surprisingly they did not appear to have much influence on rhinovirus infections, which continued to circulate widely.
“Basically, there were only two viruses spreading during the pandemic restrictions, SARS-CoV-2 and rhinovirus, and we were curious about what was going on: Was one rhinovirus species dominating or was there a new, emergent variant causing these infections?” said Stephanie Goya , a postdoctoral research scientist in the Greninger lab. She was lead author of the paper about the findings.
By comparison, the relaxed restrictions of late 2022 led to a resurgence of SARS-CoV-2, influenza and RSV infections, which became known as the “tripledemic.” During this period, too, rhinovirus was found to be widely prevalent.
To find out which viral variants were circulating in the region, the researchers sequenced more than 1,000 RNA genomes of rhinovirus they detected on samples from the tens of thousands of swabs collected during the two spans.
These sequences provided a kind of genetic fingerprint, called a genotype, that allowed the researchers to determine which rhinovirus types were circulating at a given time, and to create the equivalent of family trees to work out how different genotypes were related to each other and how they evolved.
They found that no single dominant rhinovirus variant was causing infections. Instead, 99 different genotypes were circulating in the region during the study spans.
Sixty-six percent of the people whose swabs tested positive for rhinovirus reported symptoms such as sore throat, runny nose and cough. Swabs from people with symptoms tended to have more virus, but no rhinovirus species or genotype seemed to cause more symptoms than another.
The mix of circulating genotypes varied: Some were more prevalent than others at different times. Of particular interest was the finding that genotypes that predominated in the first collection period were largely replaced by other genotypes in the second collection period.
This may be evidence that immunity in the population generated in 2021 influenced which genotypes could spread the following year, Goya noted.
Analysis of the genotypes’ family trees revealed that many of these genotypes aren't new. They have circulated for up to 40 years, although each genotype has a strikingly stable protein sequence. Researchers believe that the virus’ success comes from having a variety of genotypes present at the same time rather than from a series of new evolutionary changes occurring in each of them. Because each genotype is recognized as distinct by the immune system, each genotype spreads effectively at different times.
Some changes were found in segments of the viral genome that affect the structure of the protein that latches onto cells in the nose and throat. These proteins are often important targets for antibodies and other mechanisms of immune response. Because the structural changes are few, it might be possible to create a vaccine that generates an effective immune response to large numbers of genotypes and, hopefully, prevent many common colds, Goya said.
Written by Michael McCarthy
For details about UW Medicine, please visit http://uwmedicine.org/about .
Loading metrics
Open Access
Peer-reviewed
Research Article
The assembly of neutrophil inflammasomes during COVID-19 is mediated by type I interferons
Roles Conceptualization, Data curation, Formal analysis, Investigation, Software, Validation, Visualization, Writing – original draft
* E-mail: [email protected]
Affiliation Viral Zoonosis Research Unit, Medicum, Department of Virology, University of Helsinki, Helsinki, Finland
Roles Investigation, Writing – review & editing
Affiliations Department of Bacteriology and Immunology, University of Helsinki, Helsinki, Finland, Translational Immunology Research Program, Faculty of Medicine, University of Helsinki, Helsinki, Finland
Roles Investigation
Affiliations Viral Zoonosis Research Unit, Medicum, Department of Virology, University of Helsinki, Helsinki, Finland, Department of Veterinary Biosciences, University of Helsinki, Helsinki, Finland
Roles Resources
Affiliations Viral Zoonosis Research Unit, Medicum, Department of Virology, University of Helsinki, Helsinki, Finland, Department of Veterinary Biosciences, University of Helsinki, Helsinki, Finland, Department of Tropical Parasitology, Institute of Maritime and Tropical Medicine, Medical University of Gdansk, Gdynia, Poland
Affiliations Human Microbiome Research Program, Faculty of Medicine, University of Helsinki, Helsinki, Finland, Meilahti Vaccine Research Center MeVac, Department of Infectious Diseases, Inflammation Center, Helsinki University Hospital and University of Helsinki, Helsinki, Finland
Roles Funding acquisition, Resources, Writing – review & editing
Roles Supervision, Writing – review & editing
Affiliations Department of Bacteriology and Immunology, University of Helsinki, Helsinki, Finland, Translational Immunology Research Program, Faculty of Medicine, University of Helsinki, Helsinki, Finland, Division of Virology and Immunology, HUSLAB Clinical Microbiology, HUS Diagnostic Center, Helsinki University Hospital, Helsinki, Finland
Affiliation Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland
Roles Resources, Writing – review & editing
Roles Investigation, Methodology, Supervision, Validation, Visualization, Writing – review & editing
Affiliations Department of Veterinary Biosciences, University of Helsinki, Helsinki, Finland, Laboratory for Animal Model Pathology, Institute of Veterinary Pathology, Vetsuisse Faculty, University of Zurich, Zurich, Switzerland, Department of Infection Biology & Microbiomes, Institute of Infection, Veterinary and Ecological Sciences, University of Liverpool, Liverpool, United Kingdom
Roles Funding acquisition, Resources
Affiliations Viral Zoonosis Research Unit, Medicum, Department of Virology, University of Helsinki, Helsinki, Finland, Department of Veterinary Biosciences, University of Helsinki, Helsinki, Finland, Division of Virology and Immunology, HUSLAB Clinical Microbiology, HUS Diagnostic Center, Helsinki University Hospital, Helsinki, Finland
Roles Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Supervision, Validation, Writing – original draft
- Luz E. Cabrera,
- Suvi T. Jokiranta,
- Sanna Mäki,
- Simo Miettinen,
- Ravi Kant,
- Lauri Kareinen,
- Tarja Sironen,
- Jukka-Pekka Pietilä,
- Anu Kantele,

- Published: August 22, 2024
- https://doi.org/10.1371/journal.ppat.1012368
- Peer Review
- Reader Comments
The severity of COVID-19 is linked to excessive inflammation. Neutrophils represent a critical arm of the innate immune response and are major mediators of inflammation, but their role in COVID-19 pathophysiology remains poorly understood. We conducted transcriptomic profiling of neutrophils obtained from patients with mild and severe COVID-19, as well as from SARS-CoV-2 infected mice, in comparison to non-infected healthy controls. In addition, we investigated the inflammasome formation potential in neutrophils from patients and mice upon SARS-CoV-2 infection. Transcriptomic analysis of polymorphonuclear cells (PMNs), consisting mainly of mature neutrophils, revealed a striking type I interferon (IFN-I) gene signature in severe COVID-19 patients, contrasting with mild COVID-19 and healthy controls. Notably, low-density granulocytes (LDGs) from severe COVID-19 patients exhibited an immature neutrophil phenotype and lacked this IFN-I signature. Moreover, PMNs from severe COVID-19 patients showed heightened nigericin-induced caspase1 activation, but reduced responsiveness to exogenous inflammasome priming. Furthermore, IFN-I emerged as a priming stimulus for neutrophil inflammasomes. These findings suggest a potential role for neutrophil inflammasomes in driving inflammation during severe COVID-19. Altogether, these findings open promising avenues for targeted therapeutic interventions to mitigate the pathological processes associated with the disease.
Author summary
COVID-19, caused by the SARS-CoV-2, ranges from mild “flu”-like symptoms to severe respiratory distress or even death. Neutrophils are important cells of our immune system which are strongly involved in inflammatory responses, including those occurring in COVID-19. However, despite extensive research, the precise contribution of neutrophils to the pathogenesis of COVID-19 remains elusive, and further clarification on their role is still needed. In this study, we isolated neutrophils from COVID-19 patients and healthy controls to analyze changes in their gene expression profile and inflammatory functions. These analyses revealed a distinct type I interferon (IFN-I) gene signature expressed by mature, but not immature, neutrophils from severe COVID-19 patients, which was absent in mild cases and healthy controls. Additionally, neutrophils from severe COVID-19 showed signs of increased inflammasome activation, a protein complex that contributes to inflammation by releasing inflammatory cytokines. Notably, IFN-I alone was able to promote neutrophil inflammasome formation in vitro suggesting a direct link between IFN-I response and inflammasome formation during COVID-19. Furthermore, increased neutrophil inflammasome activity was detected also in a mouse model of COVID-19. These findings suggest a potential role for neutrophils in driving excessive inflammation during severe COVID-19, and a role for IFN-I in priming the assembly of inflammasomes in these cells.
Citation: Cabrera LE, Jokiranta ST, Mäki S, Miettinen S, Kant R, Kareinen L, et al. (2024) The assembly of neutrophil inflammasomes during COVID-19 is mediated by type I interferons. PLoS Pathog 20(8): e1012368. https://doi.org/10.1371/journal.ppat.1012368
Editor: Tom Gallagher, Loyola University Chicago Stritch School of Medicine, UNITED STATES OF AMERICA
Received: February 8, 2024; Accepted: June 24, 2024; Published: August 22, 2024
Copyright: © 2024 Cabrera et al. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Data Availability: All relevant data are within the manuscript and its supporting information files. RNA-seq data are deposited at the GEO (GSE272381 and GSE271808).
Funding: This work was financed by grants by the Academy of Finland to T.S. (321809), A.K. (336439 and 335527); grants by the Helsinki University Hospital funds to O.V. (TYH 2021343); EU Horizon 2020 programme VEO (874735) to O.V.; Finnish governmental subsidy for Health Science Research (TYH 2021315) to A.K.; Paulon Säätiö to L.E.C.; Suomen Lääketieteen Säätiö to L.E.C.; Jane and Aatos Erkko foundation to O.V. The funders had no role in study design, data collection and analysis, nor decision to publish, or preparation of the manuscript.
Competing interests: The authors have declared that no competing interests exist.
Introduction
Severe COVID-19 is characterized by a dysregulated immune response with an excessive production of pro-inflammatory cytokines and chemokines. Type I interferons (IFN-I) are critical antiviral cytokines in the innate immune responses against viral infections, drawing particular attention amidst the COVID-19 pandemic [ 1 – 3 ]. While the IFN-I response helps to limit virus replication [ 3 ], its prolonged and uncontrolled activation is detrimental to the overall health of the patient [ 4 ]. As part of the pro-inflammatory response, neutrophils are rapidly recruited to the site of infection in response to SARS-CoV-2 infection [ 5 , 6 ]. Prominent neutrophil recruitment in severe COVID-19 is associated with an increased number of immature low-density granulocytes (LDGs) in the circulation [ 7 – 9 ]. The increased production and subsequent early release of immature cells from the bone marrow occurs in response to emergency myelopoiesis [ 9 ]. This process is initiated by the body to enable the recruitment of innate immune cells into the tissues and to replenish the depleted leukocyte pool, in an effort to combat viral infections including SARS-CoV-2 [ 10 ]. However, the premature release of these cells could be associated with the increased degranulation and formation of neutrophil extracellular traps (NETs) reported during SARS-CoV-2 infection, to which LDGs have a higher propensity than polymorphonuclear cells (PMN) [ 5 , 6 , 11 ].
Neutrophils are involved in several aspects of inflammatory processes, including the release of reactive oxygen species (ROS) and other pro-inflammatory mediators such as Interleukin-6 (IL-6) and IL-8. In addition, recent reports on COVID-19 highlight that neutrophils could be a major source of inflammasome derived IL-1β, which has been implicated as a substantial contributor to COVID-19 pneumonia [ 12 ]. Inflammasomes are intracellular multiprotein complexes involved in the inflammatory response. In the presence of a pathogen, antigen recognition by the immune system triggers the assembly of the inflammasome, a step known as the first signal. This is followed by the recruitment of adaptor molecules that activate NOD-like receptor (NLR) family members and the binding of the apoptosis-associated speck-like protein (ASC), finally activating the inflammasome complex [ 13 ]. The triggered assembly of this complex is known as the second signal. Studies have shown that SARS-CoV-2 infection induces significant inflammasome activation in circulating and lung-infiltrating myeloid cells, such as monocytes and neutrophils [ 14 – 17 ]. However, while the precise mechanism by which inflammasomes are activated in monocytes/macrophages is well established, less is known about molecular mechanisms of inflammasome formation in neutrophils. Thus, this study investigates the inflammasome formation in neutrophils during COVID-19 in more detail, also focusing on the different developmental stages of these cells. In addition, a recently established COVID-19 mouse model served to further explore the role of IFN-I in neutrophil inflammasome assembly.
Materials and methods
Ethics statement.
The study was approved by the Ethics Committee of the Hospital District of Helsinki and Uusimaa (HUS/853/2020, HUS/1238/2020). All volunteers gave a written informed consent, in accordance with the Declaration of Helsinki. For animal experiments, experimental procedures were approved by the Animal Experimental Board of Finland (license number ESAVI/28687/2020).
Patient population
Adult clinical patients with confirmed COVID-19 (RT-PCR positive for SARS-CoV-2) at Helsinki University Hospital (HUH) (hospitalized: n = 34; outpatients: n = 8) were enrolled in the present study. Blood samples were collected during hospitalization for the severe COVID-19 group, and after confirmation of diagnosis for the mild COVID-19 outpatient group. Samples for RNA sequencing were collected in 2020 and representing infections by the original and early SARS-CoV-2 variants, whereas samples for ex vivo culture experiments were collected in 2021–2022 likely representing infections by omicron subvariants of SARS-CoV-2. As controls, healthy blood donors were included for RNA sequencing (n = 7, age 57 ± 7, male/female 3/4) and ex vivo culturing experiments (n = 9, age 38 ± 14, male/female 4/5). For clinical correlation analysis, severe COVID-19 patients were further categorized by severity based on their need for hospitalization and oxygen supplementation, as described previously [ 7 ]. For each patient, medical history and clinical data were collected through retrospective patient record review and are presented for the severe COVID-19, hospitalized patients in Table 1 and as previously described [ 7 ]. Calprotectin was measured from serum (diluted 1:1000) by ELISA, according to the manufacturer’s protocol (calprotectin/S100A8 DuoSet kit, R&D systems).
- PPT PowerPoint slide
- PNG larger image
- TIFF original image
https://doi.org/10.1371/journal.ppat.1012368.t001
The World Health Organization (WHO) Ordinal Scale for clinical improvement is a tool designed specifically to assess and measure the progression and clinical improvement of patients [ 67 ]. COVID-19 scoring: 1 = no limitations of activity, 2 = limitations of activity, 3 = no oxygen therapy, 4 = oxygen by mask or nasal cannulae, 5 = non-invasive ventilation or high-flow oxygen, 6 = invasive mechanical ventilation without other organ support, 7 = invasive mechanical ventilation with other organ support, 8 = dead. The baseline score represents the timepoint of the first laboratory sample taken, serving as a reference point for measuring improvement and establishing a starting point for comparison. In contrast, the worst score represents the most severe or critical state of the disease. None of the patients whose samples were used for RNA-seq underwent corticosteroid treatment.
Isolation of granulocytes from human blood
Blood samples from COVID-19 patients and healthy controls (HC) were collected in EDTA vacutainer tubes and transported to the laboratory. Peripheral blood mononuclear cells (PBMCs) or polymorphonuclear cells (PMNs) were isolated from whole blood by density gradient centrifugation using either Ficoll-Paque Plus (GE Healthcare) or Polymorphprep (Axis-Shield) respectively, following standard procedures including the use of 2 mM EDTA in PBS and red blood cell lysis with ACK lysis buffer (Lonza by Thermo Fisher). Subsequently, isolation of CD66 + granulocytes (low-density granulocytes, LDGs) from the PBMC fraction was performed using the CD66abce MicroBead Kit (Miltenyi Biotec, Germany) with an MS column, according to the manufacturer’s instructions. Both the positively selected CD66 + LDGs and the isolated PMNs were then washed and counted, using a TC20 Automated Cell Counter (Bio-Rad Laboratories, Inc.) with trypan blue staining for dead cell exclusion. All described procedures in this section were done at room temperature. An aliquot of cells was lysed in Trizol reagent (Thermo Fisher Scientific, USA) and stored at –80°C for later extraction of total RNA and subsequent RNA sequencing (RNA-seq) analysis.
Caspase1 activity
Caspase1 activity was assessed in isolated cells after 2 h of culture (1 million cells/ml) using the caspase-Glo1 inflammasome assay (Promega) according to the manufacturer’s protocol, with 2.5 μM nigericin (Invivogen) treatment as the activator. The resulting luminescence was measured by a Hidex Sense microplate reader (Hidex).
As another approach, caspase1 activity in isolated cells (1 million cells/ml) was measured by the fluorescent dye FAM-FLICA (Bio-Rad Laboratories). Cells were incubated with FAM-FLICA according to manufacturer’s recommendations for 30 min in culture medium at 37°C, after which cells were washed with PBS and analyzed by LSRII cytometer (BD Biosciences). Data was acquired with BD FACSDiva version 8.0.1 (BD Biosciences) software and further analysis was performed with the FlowJo software v10 (BD Biosciences).
Soluble factor stimulation assays
Isolated granulocytes from HC and COVID-19 patients were cultured at 2 million cells/ml in RPMI 1640 supplemented with 10% fetal bovine serum (R10) at 37°C. Cells were primed (1 st signal) with either LPS (20 ng/ml, Sigma Aldrich) or IFN-I (combination of 2.7*10 4 IU/ml IFN-α and IFN-β, Immunotools) for 4 h, followed by activation (2 nd signal) by 2.5 μM nigericin or monosodium urate crystals (MSU, 100 μg/ml, Invivogen) for an additional 4 h. For the 24 h stimulation experiments, nigericin was added to the cultured cells, in the presence or absence of inflammasome inhibitors MCC995 (2 μg/ml) and Ac-YVAD-FMK (20 μg/ml, both from Invivogen). Cells were pelleted by centrifugation at 400 G for 5 min and stored in Trizol at –80°C for later RNA extraction whereas supernatants were used to measure IL-1β, IL-18, myeloperoxidase (MPO) and IL-8 by ELISAs according to the manufacturer’s protocols (DuoSet kits from R&D systems). LDH was measured in supernatants using Cyquant LDH cytotoxicity assay (ThermoFisher). Where indicated, priming and activation was performed in the presence of 100 μg/ml of anti-human IFNAR1 (Anifrolumab Biosimilar) or mouse IgG1 as control (both from Bio-X-Cell, New Hampshire, USA).
HL-60 cells (ATCC #CCL-240) were activated similarly to neutrophils after a 5-day differentiation period induced by 1% DMSO.
Virus propagation
The SARS-CoV-2 hCoV-19/Finland/THL-202117309/2021 (delta strain B.1.617.2) and the mouse-adapted strain MaVie [ 18 ] were propagated in VeroE6-TMPRSS2 cells (kidney epithelial cells expressing the transmembrane protease serine 2) [ 19 ] grown in DMEM supplemented with 10% inactivated FCS, 100 IU/mL Penicillin, 100 μg/mL Streptomycin and 2 mM L-glutamine at 37°C. The virus was purified from supernatants by ultracentrifugation (SW28 rotor, 27,000 rpm, 90 min, +4°C) through a 0.22 μm-filtered 30% ultra-pure sucrose cushion (in PBS), to obtain virus preparations free of cell culture contaminants. Virus titers were calculated by the median tissue culture infectious dose (TCID50) after assessing cytopathic effects by crystal violet staining of cell cultures infected for 5 days with serially diluted virus.
RNA sequencing
Neutrophils isolated from different cohorts comprised three PMN groups (severe COVID-19, mild COVID-19, and healthy controls), and one LDG group (given that these cells were rare in mild COVID-19 patients and HC, only LDGs from patients with severe COVID-19 were included).
cDNA synthesis from total RNA was performed according to Takara SMARTseq v4 Ultra-low input RNA kit for Sequencing user manual (Takara Bio, Mountain View, CA, USA) followed by Illumina Nextera XT Library preparation according to Illumina Nextera XT Reference Guide (Illumina, San Diego, CA, USA). UDI index setup was used for the Nextera XT libraries. Library quality check was performed using LabChip GX Touch HT High Sensitivity assay (PerkinElmer, USA) and libraries were pooled based on the concentrations acquired from the assay. The pooled libraries were quantified for sequencing using KAPA Library Quantification Kit (KAPA Biosystems, Wilmington, MA, USA) and sequenced on the Illumina NovaSeq6000 system for 200 cycles using S1 flow cell (Illumina, San Diego, CA, USA). Read length for the paired-end run was 2x101 bp. The human RNA-seq data are deposited at the GEO (accession number GSE#272381).
RNA data analysis
Principal Component Analysis (PCA) and enrichment analyses were obtained using ExpressAnalyst [ 20 ]. Briefly, PCA was performed to identify patterns in the data and reduce the dimensionality of the dataset, where the top principal components were selected based on the percentage of variance explained. For enrichment analyses, Gene Set Enrichment Analysis (GSEA) and Over-Representation Analysis (ORA) were performed on the top 5000 DE genes identified by DESeq2 (adjusted P value < 0.05, log2FC >1) [ 20 ]. GSEA was used to identify enriched signaling pathways using the Reactome database, while ORA was used to identify enriched pathways using the KEGG database. The resulting p-values were corrected for multiple testing using the Benjamini-Hochberg method, and pathways with a corrected p-value <0.05 were considered significant.
To visualize the expression patterns of the DE genes, the data was analyzed using the AltAnalyze software [ 21 ], which selected the top 118 genes based on correlation and determined the heatmap clustering, using the Euclidean distance metric and the complete linkage method. Then, the obtained heatmap was re-generated using heatmapper.ca [ 22 ] for better visualization.
CIBERSORTx, a machine learning algorithm that infers cell type proportions using a reference gene expression matrix of known cell types was used to perform RNA-seq deconvolution on the gene expression data to estimate the abundance of immune cell types in the samples [ 23 ]. The signature matrix used was taken from Lasalle et al . [ 8 ]. This reference matrix made use of a published whole-blood single-cell dataset [ 9 ], and included the main immune cell types: monocytes, NK cells, T lymphocytes, B lymphocytes, plasmablasts and neutrophils, the latter subclassified into mature and immature. The smaller subsets of granulocytes (eosinophils and basophils) are not considered separately and are most likely categorized as neutrophils in the bulk data deconvolution. Nonetheless, the resulting cell type proportions were used to compare the immune cell composition between groups.
Additionally, the determination of sample purity (>65% identified as neutrophils) served as a limiting parameter for the visualization of differentially expressed inflammasome related genes from the RNA sequencing results, which were selected and graphed in a heatmap using heatmapper.ca [ 22 ], clustered by complete linkage and ordered by Spearman’s rank.
For the GSEA of the reanalyzed RNA-seq data from LaSalle et al . [ 8 ], we used the fgsea R package using MSigDB pathway sets, as specified in S1 Table .
Volcano plots
To visualize differentially expressed (DE) genes between groups from human and mice RNA-seq results previously identified by DESeq2, a volcano plot was generated using GraphPad Prism. Genes with a P-adjusted value (padj or FDR) <0.05 were considered significant. Similarly, RNA sequencing data from GSE93996 [ 24 ] was reanalyzed, and all DE genes in ATRA-differentiated HL-60 cells were visualized in a volcano plot.
Single cell transcriptomics data analysis
This study made use of the “COVID-19 Immune Atlas: integration of 5 public COVID-19 PBMC single-cell datasets” available online [ 25 ]. This standardized data collection contains cells from different assays (10x 3’ v2, 10x 3’ v3, 10x technology and Seq-Well) and consists of a total of 239,696 cells from the peripheral blood, 3,693 of which are neutrophils. These neutrophils were further subclassified as mature (59%) and immature (41%), based on the immune atlas predetermined cell classes. This was confirmed by a CD16b expression in mature neutrophils, and a higher CD66b expression in the immature population. This data was obtained from and analyzed in the Chan Zuckerberg CELLxGENE platform [ 25 ].
Reverse transcription and quantitative PCR (RT-qPCR) for human selected human genes
Total RNA was extracted from unstimulated or ex vivo stimulated PMNs using the Trizol reagent (Invitrogen, USA) according to the manufacturer’s protocol. Subsequently, cDNA synthesis was performed using the RevertAid RT Reverse Transcription Kit (Thermo Scientific, USA) as per the manufacturer’s instructions. Quantitative PCR (qPCR) was performed using the Stratagene model (Agilent Technologies) and SYBR Green/ROX master mix (Thermo Scientific, USA). The primer sequences for qPCR are presented in S2 Table .
Primer specificity was confirmed using melting curve analysis and dissociation curves. The relative expression levels of the genes of interest were calculated using the 2-ΔΔCT method and normalized to the expression of the housekeeping gene GAPDH. Baseline gene expressions of unstimulated samples were statistically assessed using the Mann-Whitney test, while the two-way ANOVA Tukey’s multiple comparisons test was performed for the ex vivo stimulated samples.
Mouse infections
Female BALB/c mice (Envigo, Indianapolis, IN, USA; 7 to 8 weeks, n = 36 in total) were transferred to the University of Helsinki biosafety level-3 (BSL-3) facility and acclimatized to individually ventilated biocontainment cages (ISOcage; Scanbur, Karl Sloanestran, Denmark) for 7 days with ad libitum water and food (rodent pellets). For subsequent experimental infection, the mice were placed under isoflurane anesthesia and inoculated intranasally with 50 μL of SARS-CoV-2 MaVie strain (5*10 5 TCID50/animal) or PBS (mock-infected control). Daily weighting of all mice was performed, and their well-being was carefully monitored for signs of illness (e.g., changes in posture or behavior, rough coat, apathy, ataxia). Euthanasia was performed by cervical dislocation under terminal isoflurane anesthesia. All animals were dissected immediately after euthanasia, and the lungs were sampled for multiple downstream analyses. The infections were performed as 4 separate experiments (exp): 1) Exp 1 included 8 mice infected with MaVie and 4 mock infected mice. At 2 days post infection (dpi), 4 infected and the mock infected mice were euthanized; the remaining infected mice were euthanized at 4 dpi. The right lung was sampled for virus-specific RT-qPCR (1/5) and neutrophil isolation (4/5), the left lung was fixed for histological and immunohistochemical examination. 2) Exp 2 included 8 infected and 4 mock infected mice of which half were euthanized at 2 dpi and 4 dpi, respectively. From these mice, both lung lobes were subjected to neutrophil isolation. 3) Exp 3 included 8 mice that were infected and immediately inoculated intraperitoneally with 250 μg of anti-mouse IFNAR-1 (n = 4) or mouse IgG1 control (n = 4) (Bio-X-Cell, New Hampshire, USA), and 4 mock-infected animals. All mice were euthanized at 2 dpi. Each 1/5 of the left lobe was processed for virus-specific RT-qPCR and histology/immunohistochemistry, respectively. The remaining 4/5 of the lungs served for neutrophil isolation. 4) Exp 4 included 4 animals in the following 5 groups: PBS-inoculated controls, infected animals euthanized at 2 dpi and at 4 dpi respectively, infected animals euthanized at 2 dpi with intraperitoneal injection of control IgG or anti-mouse IFNAR-1. Each 1/5 of the left lobe was processed for virus-specific RT-qPCR and histology/immunohistochemistry, respectively. The remaining lung tissue served to prepare single cell suspensions. Each 1 million cells were subjected to neutrophil quantification or caspase1 activity measurement by flow cytometry, the rest (approx. 8 million cells) to neutrophil isolation.
Virus titration from mouse lungs
Supernatants of single cell suspensions from Exp 4 were used for virus titration by fluorescent focus forming unit (FFU)-based assay, in which Vero E6 cells were incubated with serially diluted supernatants for 24 hours at 37°C in growth medium. Cells were fixed with 4% PFA for 10 min, blocked and permeabilized with PBS containing 3% BSA and 0.2% TritonX-100 for 15 min and incubated with rabbit anti- SARS-CoV-2 receptor binding domain (RBD) [ 19 ] for 1 hr, followed by anti-rabbit AlexaFluor488 conjugated secondary antibody (Thermo Scientific) for 1 hr. Fluorescence was observed with Zoe fluorescence imager (Bio-Rad laboratories) and the highest dilution not showing any RBD positive cells considered as the virus titer.
Neutrophil quantification and caspase1 activity measurement from mouse lungs by flow cytometry
Single cell suspensions from mouse lungs of Exp 4 were subjected to flow cytometric quantification of Ly6G+ neutrophils. Cells were initially incubated with BV605-conjugated yellow live/dead dye (Thermo Scientific) for 15 min before addition of 1% FBS and a cocktail of antibodies recognizing CD3+ and CD19+ lymphocytes (FITC-conjugated clones 145-2C11 and 1D3, respectively, from Immunotools), Ly6G+ neutrophils (PE-Cy7-conjugated clone 1A8 from BD biosciences) and CD11b (APC-conjugated clone M1/70.15 from Immunotools). After incubation for 30 min at RT, cells were fixed with 2% paraformaldehyde for 30 min and washed with PBS. In parallel, single cell suspensions were stained with FAM-FLICA together with Ly6G antibody and 30 min incubations were performed in R10 at 37°C before fixation. Finally, all cells were subjected to flow cytometric analysis with a three-laser (Blue/Red/Violet lasers) 14-color Fortessa LSRII cytometer (BD Biosciences). Data was acquired with BD FACSDiva version 8.0.1 (BD Biosciences) software and further analysis was performed with the FlowJo software v10 (BD Biosciences).
Neutrophil isolation from mouse lungs
Neutrophil isolation was performed from the lungs of all mice. The dissected lung tissue was chopped into small pieces using scissors and enzymatically digested with a cocktail of Liberase (50 ug/ml; Roche #05401020001 from Merck) and DnaseI (100 ug/ml; Roche #11284932001 from Merck) in RPMI-1640 for 30 min at 37°C. The resulting homogenate was diluted 10-fold in R10 and passed through a 70 μm Cell strainer (Pluriselect) to obtain a single-cell suspension. Neutrophils were isolated by positive selection using Ly6G-binding magnetic beads and MS columns according to the manufacturer’s recommendations (Miltenyi Biotec). Neutrophils were isolated with a purity exceeding 95% based on flow cytometry analysis of Ly6G expression. Where indicated, isolated neutrophils were attached to glass slides through cytospin (800 g, 5 min), fixed with 4% PFA for 10 min and the nuclei stained with Hoechst33342.
RNA sequencing of mouse neutrophils
Mouse neutrophils from Exp 1 were isolated, lysed in Trizol (Thermo Scientific) and the RNA extracted in the liquid phase using chloroform. RNA isolation was carried out using the Rneasy micro kit (Qiagen). Isolated RNA (1 ng) underwent whole transcriptome sequencing with ribodepletion. Briefly, RNA sequencing was performed using the Illumina Stranded with RiboZero library preparation method. Sample quality and integrity were assessed using TapeStation RNA analysis. Sequencing was conducted on the Illumina NextSeq platform, followed by standard bioinformatics analysis for gene expression quantification.
The service was provided by the Biomedicum Functional Genomics Unit at the Helsinki Institute of Life Science and Biocenter Finland at the University of Helsinki. The mouse RNA-seq data are deposited at the GEO (GSE271808).
RT-qPCR of mouse samples
RNA was extracted from dissected lung samples (1/10 of the whole lung) as well as isolated neutrophils of mice in Exp 1, 3 and 4 using Trizol (Thermo Scientific) following the manufacturers’ instructions. The isolated RNA was directly subjected to one-step RT-qPCR analysis based on a previously described protocol using primer-probe sets detecting the viral genome encoding for the RNA-dependent RNA polymerase (RdRp [ 26 ], subgenomic E [ 27 ] as well as mouse OasL2, caspase1, IL1b and GAPDH (Applied biosystems #Mm01336189_m1, #Mm00438023_m1, #Mm00434228_m1 and #Mm99999915_g1 respectively, Thermo Scientific). The PCRs were performed with TaqPath 1-step master mix (Thermo Scientific) using AriaMx instrumentation (Agilent, Santa Clara, CA, USA).
Histology and immunohistochemistry
From animals in Exp 1, 3 and 4 the whole left lung (Exp 1) or 1/5 of the left lung (Exp 3 and 4) were trimmed for histological examination and routinely paraffin wax embedded. Consecutive sections (3 μm) were prepared and routinely stained with hematoxylin-eosin (HE) or subjected to immunohistochemistry (IHC) for the detection of SARS-CoV-2 nucleoprotein (NP) [ 28 ] and Ly6G (neutrophil marker); for Exp 3, a further section of the infected lungs was stained for histone H3 (NET marker) [ 29 ]. All stains followed previously published protocols [ 30 ].
Morphometric analyses
For quantification of SARS-CoV-2 antigen expression and the extent of neutrophil influx into the lungs, a morphometric analysis was undertaken on the slides stained for SARS-CoV-2 NP and Ly6G, respectively. The stained slides were scanned using NanoZoomer 2.0-HT (Hamamatsu, Hamamatsu City, Japan), and several sections of the lung of each animal were quantitatively analysed using the Visiopharm 2022.01.3.12053 software (Visiopharm, Hoersholm, Denmark). The average total tissue area used for quantification was 19.5 ± 6 mm 2 . The morphometric analysis served to quantify the area, in all lung sections of an animal, that showed immunostaining for viral NP and Ly6G, respectively. In Visiopharm, for each section, the lung was manually outlined and annotated as a Region Of Interest (ROI), manually excluding artifactually altered areas. The manual tissue selection was further refined with an Analysis Protocol Package (APP) based on a Decision Forest classifier, with the pixels from the ROI being ultimately classified as either “Tissue” or “Background”. This new “Tissue” ROI, regrouping the different lung samples analysed for each animal, was further quantified by executing two APPs successively. The first APP was based on a Threshold classifier and served to detect and outline areas with immunostaining. The second APP then measured both the surface of the immunostained area (μm 2 ) and the surface of the “Tissue” ROI (μm 2 ). The percentage of immunostained area (%), expressed as the ratio between the immunostained area and the total area, was obtained for each animal in Excel (Microsoft Office 2019; Microsoft, Redmond, Washington, United States), according to the following formula: ([positive area (μm2)]/ [total area (μm2)]) x 100.
Statistical analyses
Statistical analysis was performed using GraphPad Prism 8.3 software (GraphPad Software, San Diego, CA, USA) and R software v3.6.3 (R core team). Statistically significant correlations between parameters were assessed by calculating Spearman’s correlation coefficients, and differences between groups were assessed with Mann-Whitney, Kruskall-Wallis or ordinary one-way or 2-way ANOVA tests, depending on sample distribution and the number of groups analyzed. To elaborate, nonparametric tests like Mann-Whitney and Kruskall-Wallis were employed when the data violated assumptions of normality, while ANOVA tests were applied when the data met parametric assumptions.
Unsupervised RNA-seq analysis reveals an antiviral gene expression signature of circulating neutrophils in COVID-19 that is strongly influenced by maturity
Different neutrophil subsets have gained a lot of attention as modulators of COVID-19 pathogenesis. We recently found increased frequencies of one neutrophil subset, referred to as low-density granulocytes (LDGs, isolated from the PBMC fraction), during COVID-19 [ 7 ]. In the current study we sought to understand in more detail the transcriptomic profile of different neutrophil subsets, including LDGs and their “normal” density counterpart, the circulating polymorphonuclear cells (PMNs) [ 31 ], which typically consists of mainly mature neutrophils. Neutrophils isolated from different cohorts comprised three PMN groups (severe COVID-19, mild COVID-19, and healthy controls), and one LDG group. Initial deconvolution of the RNA sequencing (RNA-seq) data allowed us to gain a comprehensive understanding of the cellular composition within PMN and LDG fractions and verified that most cells present in the samples were neutrophils ( S1A Fig ). This analysis also demonstrated that cells in the LDG fraction were predominantly immature neutrophils, meanwhile PMNs were composed of mainly mature neutrophils.
The samples with predominant neutrophil cell populations were selected for subsequent gene expression analysis (neutrophils ≥ 65%). The high variance in gene expression between PMNs and LDGs was confirmed by principal component analysis (PCA) ( Fig 1A ), which revealed that the gene expression patterns of COVID-19 LDGs differed from those of all PMNs regardless of the patients’ disease state. Functional enrichment analyses through gene overrepresentation (ORA) and gene-set enrichment analyses (GSEA) ( Fig 1B ) compared PMNs with LDGs from severe COVID-19 patients. The most statistically significant result was an overrepresentation of the NOD-like receptor signaling pathway in PMNs in contrast with LDGs, highlighting that the different neutrophil fractions have a distinct inflammatory profile. This was supported by GSEA, where the most obvious increases in fold changes were the enrichment of the interferon signaling pathways. Another relevant difference was the cell cycle and DNA replication pathways, identified by both ORA and GSEA, which supported our previous findings suggesting LDGs to be predominantly immature cells [ 7 ]. Furthermore, a heatmap of selected type I IFN (IFN-I) related genes confirmed a robust IFN-I gene signature in severe COVID-19 PMNs, while LDGs from severe COVID-19 distinctively lacked this signature ( Fig 1C ). Unsupervised clustering analysis, namely Iterative Clustering and Guide Gene Selection (ICGS) using the AltAnalyze software, supported these findings by identifying the top 118 differentially expressed (DE) genes, including several IFN-related genes ( S1B Fig ). Similarly to the selected samples included in Fig 1 , this analysis classified the samples into two major clusters: a first one containing all isolated LDG samples, and a second one comprising all isolated PMN samples. The former cluster consisted of neutrophil antimicrobial and granule marker genes (e.g. DEFA3 , DEFA4 , SERPINB10 , CTSG ), while in the latter cluster the most significantly upregulated genes in the PMNs from severe COVID-19 subgroup were mainly interferon inducible (e.g. IFI44L , IFI6 , GBP3 , IRF7 ). These differences were supported by a detailed gene analysis ( S2A Fig ).
The analysis was reduced to include only the samples with the highest purity (cell fraction over 0.65 of neutrophils), as identified by CIBERSORTx. ( A ) Principal component analysis (PCA) of the RNA-seq samples (n = 7 PMNs from HC, n = 10 PMNs from severe COVID-19, n = 8 PMNs from mild COVID-19, and n = 6 LDGs from severe COVID-19). ( B ) Ridgeline diagrams depicting the top 20 enriched signal pathways from the genes differentially expressed by PMNs versus LDGs during severe COVID-19: overrepresentation analysis (ORA) using KEGG database and gene-set enrichment analysis (GSEA) according to Reactome database. Both enrichment analyses were made using ExpressAnalyst and are sorted by P-value, obtained from Welch’s t-test. ( C ) Heatmap of differentially expressed IFN-related genes in COVID-19 PMNs and LDGs as compared to HC PMNs. RNA sequencing was performed on purified PMNs from healthy controls, mild COVID-19 and severe COVID-19, as well as LDGs from severe COVID-19. The heatmap was clustered by complete linkage and ordered by Spearman’s rank. FC = fold change .
https://doi.org/10.1371/journal.ppat.1012368.g001
Increased expression of inflammasome related genes in severe COVID-19 PMNs
In addition to the strong IFN-I signature, PMNs of severe COVID-19 upregulated several genes involved in inflammatory processes, such as the formation of inflammasomes. We further analyzed the differential expression of selected inflammasome related genes across all samples using RNA-seq ( Fig 2A ). PMNs of severe COVID-19 displayed higher levels of inflammasome genes such as NLRP3 and caspases 1, 4 and 5. LDGs did not display similar upregulation of inflammasome genes, with the notable exception IL-18 and NLRC4, which were not upregulated by PMNs. These findings prompted us to look more closely into PMN fractions between different disease states. Pathway analyses identified the inflammasome related NOD-like and RIG-like receptor signaling pathways among the most significantly overrepresented pathways, differentially expressed in severe COVID-19 PMNs versus HC PMNs (Figs 2B and S2B and S2C ) or mild COVID-19 PMNs (Figs 2C and S2D and S2E ). However, mild COVID-19 PMNs did not significantly differ from HC PMNs in their inflammatory profile ( S2F Fig ). The increased expression of selected IFN-I ( OAS1 , OAS2 , and IFIT1 ) and inflammasome related genes ( CASP1 , CASP5 , NLRC5 and NAIP ) between COVID-19 and HC PMNs was confirmed by RT-qPCR. However, some inflammasome related genes ( IL-1β , NLRP3 and NLRC4 ) were seemingly downregulated, although not statistically significant ( S3 Fig ).
( A ) Heatmap depicting selected differentially expressed inflammasome related genes from RNA sequencing performed in PMNs from HC, mild and severe COVID-19, as well as severe COVID-19 LDGs. Only the samples with the highest purity, determined by a cell fraction over 0.65 of neutrophils (identified by CIBERSORTx) are included. The heatmap was clustered by complete linkage and ordered by Spearman’s rank. ( B-C ) Ridgeline diagrams of overrepresentation analyses (ORA) according to KEGG database, depicting the top 10 enriched signaling pathways in PMNs during severe COVID-19 compared to ( B ) healthy controls and ( C ) mild COVID-19. ( D ) UMAP analysis of the COVID-19 Immune Atlas, which integrates 5 public COVID-19 PBMC single-cell transcriptomics datasets, created using CELLxGENE. (Top) UMAP showing the clustering of CD16+ cells (mature, FCGR3B expressing cells) and CD66b+ cells (immature, CEACAM8 expressing cells). Each dot represents a single cell colored according to the expression level of a selected gene. The color scale ranges from green (low expression) to purple (high expression). (Bottom) Pie chart summarizing the percentage of mature (black) and immature (blue) cells in the data. ( E ) The fraction of mature and immature neutrophils cells expressing inflammasome related genes identified in ( D ) are shown in bar graphs. For each gene, the proportion of expressing cells is shown in light blue, while the proportion of negative or not-expressing cells is shown in gray. Zoomed-in bar graph depicts the proportion of mature and immature cells expressing each gene.
https://doi.org/10.1371/journal.ppat.1012368.g002
Single cell sequencing data from the COVID-19 immune atlas, which integrates data from 5 independent studies analyzing COVID-19 PBMCs, confirmed our transcriptomic results ( Fig 2D ), from which a detailed gene by gene analysis of the most relevant inflammasome related genes is shown (Figs 2E and S4 ). Briefly, PYCARD gene coding for the ASC protein was expressed similarly in mature and immature neutrophils ( S4 Fig ), suggesting that both cell types may have ASC-dependent inflammasome forming capacity. However, most of the inflammasome gene expressions differed significantly and in the same manner as in our transcriptomic analysis.
Activation of neutrophil inflammasome related pathways during respiratory distress is not specific to COVID-19
We also reanalyzed the RNA-seq data generated by LaSalle et al . [ 8 ], focusing on neutrophil transcriptomics in patients with COVID-19 versus non-COVID-19 patients, and healthy controls. The non-COVID-19 patients exhibited acute respiratory distress and clinical suspicion for COVID-19. However, they tested negative for SARS-CoV-2 by PCR, unlike those classified as COVID-19 patients. Our analysis included IFN-α response, IL-1β production, TLR signaling, NLRP3 inflammasome, and pyroptosis pathways, using the Gene Ontology (GO) database; the NLR signaling pathway using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database; and inflammasome pathway using the REACTOME database ( Fig 3 ). These pathways were significantly enriched in COVID-19 patients, supporting our findings. Importantly, the genes from the above-mentioned pathways were also induced in non-COVID-19 patients, suggesting that these pathways represent a general neutrophil response to inflammatory stimuli rather than a COVID-19 specific response.
Bar graphs represent the activation levels of selected pathways and processes as identified by neutrophil transcriptomics. The analysis includes interpheron alpha (IFN-α) responses, interleukin (IL)-1β production, Toll-like receptor (TLR) signaling, NLRP3 inflammasome, and pyroptosis, as determined through the Gene Ontology (GO) database. The NOD-like receptor signaling pathway was investigated using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database, and the inflammasome pathway was explored via the REACTOME database (more information in S1 Table ). The graphs compare the activation levels of these pathways in healthy controls (HC), non-COVID patients with similar symptoms (COVID-19 negative), and COVID-19 positive individuals. Statistical significance is denoted as follows: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. P values were calculated with Kruskall-Wallis test.
https://doi.org/10.1371/journal.ppat.1012368.g003
Inflammasomes are activated in severe COVID-19 PMNs, but not directly by SARS-CoV-2
Given the strong upregulation of many inflammasome related genes during severe COVID-19, we assessed whether PMNs exhibit active inflammasome formation in vivo . To evaluate spontaneous inflammasome mediated cytokine secretion, fresh PMNs isolated from severe COVID-19 patients and HC were cultured ex vivo for 24 hours. We measured the levels of IL-1β and IL-18 in the supernatant and found that IL-1β secretion was significantly increased in the supernatant of severe COVID-19 PMNs compared to HC PMNs ( Fig 4A ), whereas the IL-18 levels did not differ significantly ( Fig 4B ). Additionally, since SARS-CoV-2 viral particles were previously implicated to induce inflammasome formation in macrophages [ 17 ], the IL-1β and IL-18 levels after HC PMNs exposure to SARS-CoV-2 were also assessed but no significant effects in the secretion of these cytokines were observed ( Fig 4C and 4D ).
PMNs were freshly isolated from blood and cultured at 2 million cells/ml ( A ) IL-1β and ( B ) IL-18 levels in 24 h cell culture supernatants from COVID-19 (n = 11 for IL-1β and 9 for IL-18) and HC PMNs (n = 6 for both). ( C ) IL-1β and ( D ) IL-18 levels in 24 h cell culture supernatant from PMNs exposed or non-exposed to purified SARS-CoV-2 viral particles (10 virus particles / PMN) (n = 3). ( E ) Caspase1 activity in PMNs following a 2 h stimulation with nigericin or purified SARS-CoV-2 viral particles (10 virus particles / PMN). For HC PMNs, n = 9 for mock and nigericin and n = 6 for SARS-CoV-2 exposure. For COVID-19 PMNs, n = 12 for mock and nigericin and n = 9 for SARS-CoV-2 exposure. *p < 0.05 and **p < 0.01. Data presented as mean ± SD. Tukey’s multiple comparisons test for mixed-effect analysis was applied for ( E ), meanwhile P values for ( A-D ) were calculated with the Mann-Whitney U-test.
https://doi.org/10.1371/journal.ppat.1012368.g004
The spontaneous secretion of IL-1β by COVID-19 PMNs suggests that these cells are actively producing and releasing IL-1β through inflammasome formation which is dependent on caspase1 activity [ 32 ]. We assessed caspase1 activity in response to the second signal required for inflammasome activation, induced by nigericin, and observed increased caspase1 activity in severe COVID-19 PMNs compared to HC PMNs ( Fig 4E ). These findings suggest that severe COVID-19 PMNs have an increased capacity for inflammasome activation, potentially due to an existing priming signal during acute disease in vivo . However, no significant difference in caspase1 activity between non-exposed and virus-exposed PMNs were observed ( Fig 4F ), indicating that caspase1 activation in COVID-19 PMNs is not directly triggered by the virus.
Type I IFNs prime PMNs for inflammasome activation
Since PMNs from COVID-19 patients concomitantly display a strong IFN-I signature ( Fig 1B and 1C ) and an increased propensity for inflammasome activation, we hypothesized that IFN-I could act as the priming signal for PMN inflammasomes during COVID-19. Isolated HC PMNs were stimulated ex vivo with exogenous IFN-I and the well-described inflammasome priming (1 st signal) and activator (2 nd signal) agents LPS and nigericin, respectively [ 33 , 34 ]. After stimulation, both priming signals induced pro-IL-1β (31 kDa) in the cell lysates, followed by the release of active IL-1β (17 kDa) into the supernatant in response to nigericin ( Fig 5A ), confirming the ability of IFN-I to prime PMNs for inflammasome activation.
Isolated HC or COVID-19 PMNs were non-stimulated or stimulated 4h with IFN-I (combination of 2.7*10 4 IU/ml IFN-α and IFN-β) or 20 ng/ml LPS (1 st signal), followed by 4h with 2.5 μM nigericin or purified SARS-CoV-2 (10,1 virus/PMNs) (2 nd signal). Then, ( A ) western blot of pro-IL-1β (31 kD) and active IL-1β (17 kD) was performed from HC PMNs supernatant and cell lysates, ( B ) IL-1β (n = 5 HC PMN and 9 COVID-19 PMN) and ( C ) MPO (n = 5 HC PMN and 9 COVID-19 PMN) were measured from supernatants by ELISA. ( D-E ) Effect of inflammasome inhibitor MCC950 (2 μg/ml, added simultaneously with nigericin) on IL-1β secretion in ( D ) HC and ( E ) severe COVID-19 PMN supernatant (n = 3). ( F ) LDH and ( G ) IL-8 in HC and severe COVID-19 PMN supernatants (n = 3). ( H-K ) RT-qPCR of selected mRNAs in IFN-I or LPS-primed HC and COVID-19 PMNs (n = 6–8 HC PMN and 7–10 COVID-19 PMN). ( L-M ) HC PMNs were stimulated with high dose IFN-I (2.7*10 5 IU/ml), normal dose IFN-I (2.7*10 4 IU/ml) and 20 ng/ml LPS. After 4 hr stimulation caspase1 activity was measured using median fluorescence intensity (MFI) of FAM-FLICA by flow cytometry ( L , n = 5, representative histogram of one donor shown) and after 24 hr stimulation IL-1β release was measured by ELISA ( M, n = 5 ) . P values calculated with Kruskall-Wallis test for the comparison between treatments by group (HC or COVID-19 PMNs), and Mann-Whitney test for the comparison between HC and COVID-19 PMNs by individual treatment for ( B-G ), and Two-way ANOVA Tukey’s multiple comparisons test for ( B, H-K ). The treatments in L-M were compared to mock by one-way ANOVA for repeated measures, corrected for multiple comparisons with the two-stage step-up method of Benjamini, Krieger and Yekutieli. *p < 0.05, **p < 0.01, ***p < 0.001, **** p < 0.0001. Data presented as mean ± SD.
https://doi.org/10.1371/journal.ppat.1012368.g005
To assess inflammasome formation in circulating neutrophils during COVID-19, PMNs from HC and COVID-19 patients underwent similar stimulation assays as above, followed by IL-1β measurement from supernatants by ELISA. In addition, to further assess the role of SARS-CoV-2 virus particles in neutrophil inflammasome activation, HC PMNs were cultured in the presence of purified viruses (10 infectious units/PMN). HC PMNs responded to both LPS and IFN-I by increasing their IL-1β secretion, which was exponentiated after exposure to nigericin ( Fig 5A and 5B ), confirming the ability of IFN-Is to prime for inflammasome assembly in PMNs, albeit less efficiently than LPS. Furthermore, as expected, the ability of IFN-I to prime for nigericin-mediated inflammasome activation was dependent on the IFN-I receptor IFNAR1 for both IFN-α and IFN-β ( S5A Fig ).
Interestingly, COVID-19 PMNs produced less IL-1β than HC PMNs upon exogenous inflammasome activation primed by either LPS or IFN-I, while SARS-CoV-2 particles did not have any effect on PMN inflammasome activation ( Fig 5B ). As with 24 h cultures ( Fig 4B ), we did not detect any significant changes in IL-18 secretion in either HC or COVID-19 PMNs ( S5B Fig ). However, the release of myeloperoxidase (MPO), used as a marker of degranulation and/or NETosis, in response to nigericin was similar between COVID-19 PMNs and HC PMNs ( Fig 5C ), and therefore the observed diminished IL-1β release by COVID-19 PMNs is not due to general cellular inertia but may be specific to the ex vivo induced inflammasome pathway. Furthermore, additional stimulation assays in the presence of the NLRP3 inhibitor MCC950 ( Fig 5D and 5E ) and caspase1 inhibitor YVAD ( S5C Fig ) confirmed that induced IL-1β secretion is dependent on canonical NLRP3 inflammasome activation. Unlike IL-1β ( S5D Fig ), increased IL-18 secretion was not detectable even after 24 h stimulation ( S5E Fig ). Furthermore, the observed residual IL-18 was not affected by inflammasome inhibitors, suggesting its secretion to be unrelated to inflammasome activity in PMNs.
We further assessed the specificity of inflammasome activation by measuring LDH and IL-8 levels in the supernatants from the same cells and under the same experimental conditions as shown in Fig 5D and 5E . The measurements of the former were done to assess inflammasome mediated cell death by pyroptosis in response to nigericin, while the latter was assessed to demonstrate the responsiveness of PMNs to an inflammasome unrelated inflammatory cascade. As with IL-1β secretion, COVID-19 PMNs were less responsive than HC PMNs to nigericin- and LPS-mediated LDH ( Fig 5F ) and IL-8 ( Fig 5G ) release, respectively. This suggests that COVID-19 PMNs are generally poorly responsive to inflammatory stimuli.
To examine this reduced responsiveness to external inflammatory priming, we evaluated the inflammasome related gene expression following ex vivo stimulation with IFN-I or LPS (Figs 5H–5K and S5F–S5I ). OAS1 gene, an interferon stimulated gene (ISG), showed significant upregulation by IFN-I in COVID-19 PMNs as compared to HC PMNs ( Fig 5H ), while the inflammasome related genes IL-1β ( Fig 5I ), CASP1 ( Fig 5J ) and NLRC5 ( Fig 5K ) were more efficiently induced in HC PMNs than COVID-19 PMNs. This suggests that the inflammasome defect in COVID-19 PMNs is at the transcriptional level when using IFN-I as the priming factor, while high OAS1 gene expression indicates transcriptional defect is restricted to individual genes.
Next, we investigated whether IFN-I could also activate caspase1 directly without the 2 nd signal to boost inflammasome activation. In addition to treating HC PMNs with IFN-I and LPS as in previous experiments, we also included another group with a higher dose of IFN-I (ten-fold) and assessed caspase1 activity after 4 hours with FAM-FLICA, a fluorescent caspase1 reactive dye ( Fig 5L ) as well as IL-1β release after 24 hours by ELISA ( Fig 5M ). Results showed that similar to LPS, high-dose IFN-I induced significant caspase1 activity, which was not observed with the normal IFN-I concentration. Despite this, the normal IFN-I concentration still resulted in increased IL-1β levels in the supernatant. Taken together, these findings suggest that the 2 nd signal is not essential for inflammasome activation by IFN-I and that IL-1β release is a more sensitive method of detecting inflammasome activity as compared to caspase1 activity in our assay setup. Thus, these results can also explain our previous observation of increased spontaneous release of IL-1β by COVID-19 PMNs even though significant caspase1 activity is only detected after nigericin-mediated boosting in vitro ( Fig 4A and 4E ).
Association between ex vivo inflammasome activation and disease severity
Our analysis of the association between ex vivo inflammasome activation (caspase1 activity and IL-1β release) and clinical markers of disease severity, including neutrophil responses, revealed intriguing links. Calprotectin is a marker of neutrophil activation or death [ 35 ] but also potentially activates the inflammasome [ 36 ]. A significant positive correlation between calprotectin plasma levels and PMN caspase1 activity ( Fig 6A and 6B ) underscores this latter possibility and highlights the interplay between inflammation and inflammasome activation in PMNs of COVID-19 patients. Furthermore, the negative association of PMN IL-1β levels (after ex vivo stimulation with IFN and nigericin) with disease severity (WHO ordinal scale, Fig 6A ) and patient neutrophil counts ( Fig 6A and 6C ) supports the exhaustion hypothesis, wherein PMNs from severe COVID-19 patients may be less responsive to stimuli due to prior in vivo activation. While these findings provide intriguing insights into the complex interplay between calprotectin release, caspase1 activity, and inflammasome activation in COVID-19, additional research is required to further elucidate these connections.
( A ) Spearman’s correlation matrix depicting the relationships among clinical parameters and results of ex vivo experimentation. For the WHO ordinal scale, the baseline parameters were used. ( B-C ) Linear regression analysis demonstrating the associations between: ( B ) Positive association between PMN Caspase1 activity, measured after ex vivo nigericin stimulation, and the levels of Calprotectin in the matched patient’s peripheral blood; ( C ) Negative association between e x vivo stimulated PMN IL-1β levels (IFN+Nig) and the blood neutrophil count in matched patients at the time of sampling (n = 12). LOS = length of stay . WHO = World Health Organization . Min = minimum . Casp1 = caspase1 . LPS or IFN + nig = lipopolysaccharide or type I interferon + nigericin ex vivo stimulation .
https://doi.org/10.1371/journal.ppat.1012368.g006
LDGs differ from PMNs in their ability to release IL-18 upon inflammasome activation
Transcriptomic analysis presented above revealed a distinct lack of IFN-I responsive and inflammasome related gene expression in LDGs as compared to PMNs of severe COVID-19 patients. This suggested that inflammasomes are not similarly regulated in LDGs as compared to PMNs during COVID-19. To assess the inflammasome forming capacity of LDGs, we conducted ex vivo stimulation assays using LDGs isolated from COVID-19 patients, similar to the approach used for PMNs described earlier. Like PMNs, IL-1β secretion by LDGs was elevated in the presence of a priming signal (IFN-I or LPS), which exponentially increased when the inflammasome activation signaling molecule nigericin was added ( Fig 7A ). Contrary to PMNs and in line with the transcriptomics data, an increased IL-18 secretion was detected ( Fig 7B ). Additionally, the secretion of both ILs by LDGs was inhibited in the presence of inflammasome specific inhibitors MCC950 and YVAD ( Fig 7A and 7B ).
These findings suggested that IFN-I can prime for inflammasome activation also in LDGs. Furthermore, the ability to release IL-18 upon neutrophil inflammasome activation varies based on cellular maturation state. To explore this further, we conducted in vitro stimulation studies using differentiated HL-60 cells, an immature neutrophil-like model [ 37 ]. Similar to LDGs from COVID-19 patients, HL-60 displayed comparable IL-18 secretion pattern upon LPS or IFN-I stimulation and nigericin-induced activation. Notably, their IL-1β release was only detected with LPS priming ( Fig 7C and 7D ). Furthermore, transcriptomic analysis revealed an upregulation of inflammasome related genes upon differentiation ( Fig 7E ). Overall, these findings suggest that neutrophils may lose the ability to secrete IL-18 in response to inflammasome activity during maturation, and increased release of neutrophil-derived IL-18 occurs primarily in disease states associated with extensive granulopoiesis and increased immature granulocyte counts in the blood, like COVID-19 [ 38 ].
( A-D ) Isolated COVID-19 LDGs or HL-60 cells (differentiated for 5 days with 1% DMSO) were non-stimulated or stimulated 4h with IFN-I or LPS (1 st signal), followed by 4h with nigericin (2 nd signal) in the presence or absence of inflammasome inhibitors MCC950 or YVAD as previously. Secretion of ( A, C ) IL-1β and ( B, D ) IL-18 were measured from the supernatants by ELISA (n = 2 for LDGs and 3–5 for HL-60). *p < 0.05 and **p < 0.01. P values calculated with Kruskal-Wallis test. Data presented as mean ± SD. ( E ) Volcano plot of differentiated vs undifferentiated HL-60 cells gene expression from GSE93996, with inflammasome related genes marked in blue. Only significant DE genes are shown (adjusted p value < 0.05).
https://doi.org/10.1371/journal.ppat.1012368.g007
Neutrophils are recruited to the lungs in SARS-CoV-2 infected mice
Hamsters and human ACE2 expressing mice infected with SARS-CoV-2 develop pulmonary inflammation including neutrophil recruitment [ 39 – 41 ]. To further assess the role of neutrophils in COVID-19, we utilized a recently developed SARS-CoV-2 mouse model [ 18 ]. This model employs the MaVie strain, serially passaged in mouse lungs and causing pneumonia like human COVID-19 in wild-type BALB-C mice [ 18 ]. Infected mice started losing weight by day 2 post-infection, with some mice reaching the clinical endpoint of 20% weight loss by day 4 ( S6A Fig , includes animals from 4 independent infection experiments, details of animal usage in S3 Table ). The first experiments were performed to study infection kinetics, histopathology and Ly-6G+ neutrophil accumulation in lungs.
Viral loads, as assessed by RT-qPCR and titration of infectious virus, were significantly higher at 2 dpi than at 4 dpi ( Fig 8A and 8B ), and viral antigen expression, widespread at 2 dpi in bronchioles and alveoli, matched this pattern (Figs 8C and S6D ). The extensive viral replication at 2 dpi was associated with degeneration of infected epithelial cells, most prominent in the respiratory epithelium, accompanied by neutrophil (Ly6G+) infiltration ( S6D Fig ). An increase in neutrophil numbers in the lungs of infected mice was observed as represented by increased Ly6G+ neutrophil/lymphocyte ratio and total Ly6G+ cell counts as assessed by flow cytometry of lung single cell suspension ( Fig 8D and 8E ; the gating strategy for Ly6G+ neutrophils is shown in S6B Fig ). A significant increase in the number of neutrophils in the lungs of infected mice compared to PBS-inoculated mice was also confirmed in situ , by morphometrical quantification of lungs sections stained for Ly6G (Figs 8F and S6D ). Detailed information on the histological and immunohistochemical features is provided in S3 Table .
Female BALB/c mice were intranasally inoculated with 5*10 5 TCID50 SARS-CoV-2 MaVie strain or PBS as control and euthanized at 2 dpi or 4 dpi. ( A ) RNA was isolated from lungs and subjected to RT-qPCR targeting viral subE and GAPDH as housekeeping gene. The relative expression of subE was measured using the comparative Ct method as compared to mock-infected control (in which subE was undetectable but set to 40 Ct) **** p < 0.0001. P values calculated with Welch’s t-test. ( B ) Infectious virus was calculated from supernatants of lung single cell suspensions of infected mice as fluorescence focus forming units (FFU) in Vero E6 cells. * p < 0.05. P values calculated with Mann-Whitney test. ( C ) Quantification based on morphometric analysis that determines the area of immunolabelling for SARS-CoV-2 nucleoprotein in relation to total tissue area. ( D ) Quantification of Ly6G neutrophil/lymphocyte ratio in lung single cell suspensions by flow cytometry. ( E ) Quantification of total Ly6G neutrophil counts in lung single cell suspensions by flow cytometry. ( F ) Quantification of Ly6G based on morphometric analysis that determines the area of immunolabelling for Ly6G in relation to total tissue area in mock-infected controls. ( G ) Quantification of median fluorescence intensity (MFI) of CD11b expression in ly6G neutrophils by flow cytometry. Representative histograms of CD11b expression in Ly6G+ neutrophils is shown. P values for C-G were calculated with ordinary one-way ANOVA using Tukey’s multiple comparison correction. Black line represents the mean. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001. ( H ) Fluorescent nuclear staining of representative magnetic-bead isolated Ly6G neutrophils by Hoechst33342. Panels A, C and F are representative of two independent experiments.
https://doi.org/10.1371/journal.ppat.1012368.g008
Furthermore, flow cytometry revealed diminished surface expression of the maturation and activation marker CD11b in Ly6G+ neutrophils of infected mice both at 2 and 4 dpi as compared to PBS controls ( Fig 8G ). However, morphologically, neutrophils from infected mice were similar to those in the PBS control mice in displaying equally multilobed nuclei (DNA staining of isolated Ly6G+ neutrophils shown in Fig 8H ; a representative flow cytometry histogram showing the purity of Ly6G+ neutrophils after isolation is provided in S6C Fig ). This suggests that the reduced expression of CD11b on the surface of neutrophils from infected mice was not the consequence of an accumulation of CD11b-negative immature neutrophils but rather an activation-related phenomenon, such as shedding or internalization.
Neutrophils from SARS-CoV-2 infected mice display IFN-I dependent caspase1 activation
Next, we investigated whether neutrophils of SARS-CoV-2 infected mice show increased caspase1 activation. We utilized the fluorescent FAM-FLICA caspase1 reactive probe in conjunction with Ly6G+ neutrophil staining to detect caspase1 activity in neutrophils by flow cytometry. Indeed, we observed significantly an increased median fluorescent intensity of FAM-FLICA in Ly6G+ neutrophils at 4 dpi, but not at 2 dpi, compared to PBS-inoculated controls ( Fig 9A ). This finding suggests increased neutrophil caspase1 activity at latter stages of the infection, concomitant with decreased viral loads.
Female BALB/c mice were intranasally inoculated with 5* 10 5 TCID50 SARS-CoV-2 MaVie strain or PBS as control. ( A ) Lungs were harvested at 2 and 4 dpi and single cell suspensions stained with FAM-FLICA and Ly6G antibody followed by flow cytometric analysis. FAM-FLICA median fluorescence intensity (MFI) was recorded in Ly6G+ neutrophils (n = 4, representative histogram image shown). * p < 0.05, ** p < 0.01. P values calculated with one-way ANOVA using Tukey’s multiple comparisons. ( B-E ) Ly-6G+ neutrophils isolated from lung single cell suspensions based on positive selection with magnetic beads. ( B - D ) RNA was isolated and subjected to transcriptomic analysis by RNA-seq. ( B ) Principal component analysis (PCA) of the PBS-inoculated control and SARS-CoV-2 infected mice lung neutrophil RNA-seq samples. ( C ) Heatmap of the top differentially expressed genes (DEGs). ( D ) Volcano plots of DEGs between neutrophils isolated from SARS-CoV-2 infected mice versus uninfected PBS-inoculated mice. Blue points represent significant terms (adjusted p-value < 0.05), while smaller gray points represent non-significant terms. Relevant inflammasome and interferon related genes are shown with larger and darker blue points. ( E ) Caspase1 activity in isolated mice neutrophils following a 2 h stimulation with nigericin was assessed by a bioluminescence method (Caspase-Glo 1 Inflammasome Assay). ( F-I ) Mice were intraperitoneally inoculated with 250 μg anti-IFNAR or IgG1 isotype control directly after infection with SARS-CoV-2 and lung neutrophils isolated at 2 dpi (including also intranasally PBS-inoculated control mice without intraperitoneal injection). ( F ) Caspase1 activity was assessed following a 2 h stimulation with nigericin by bioluminescence method. ( G-I ) RNA was isolated from isolated neutrophils and fold change mRNA expressions of ( G ) Oasl2, ( H ) Caspase1 (Casp1) and ( I ) IL-1β (Il1b) was assessed by RT-qPCR in isotype control and anti-IFNAR treated infected mice as compared to mock-infected control mice. *p < 0.05, **p < 0.01 and ***p < 0.001. P values for A, E and F panels were calculated with ordinary one-way ANOVA using Tukey’s multiple comparisons correction, while Welch’s t-test was used for panels G-I. Data presented as mean ± SD.
https://doi.org/10.1371/journal.ppat.1012368.g009
The potential role of IFN-I in mediating caspase1 activity in neutrophils of infected mice was initially assessed by isolating neutrophils from the lungs of infected mice at 2 and 4 dpi, as well as from non-infected mice, for transcriptomic analysis by RNA-seq. PCA showed differences between neutrophils from infected and non-infected mice, with slight variation between the 2 and 4 dpi time points ( Fig 9B ). These differences were reflected in many DEGs, including several IFN-I responsive and inflammasome related genes, which showed strong upregulation at 2 dpi with slightly lower but still significantly elevated levels at 4 dpi, compared to non-infected mice (highlighted in the DEG heatmap; Fig 9C ). The volcano plot ( Fig 9D ) provided a comprehensive view of the DEG pattern between neutrophils from SARS-CoV-2 infected and mock-infected mice. In addition to confirming the upregulation of IFN-I responsive and inflammasome related genes observed in the heatmap, the plot revealed a broader transcriptional response to viral infection with several additional DEG.
Having established a robust IFN-I transcriptional signature in neutrophils from SARS-CoV-2 infected mice we wanted to assess whether they possess increased propensity for nigericin-induced caspase1 activation, similar to human PMNs after exogenous priming by IFN-I. Neutrophils from infected mice, harvested at 2 and 4 dpi, displayed increased caspase1 activity upon nigericin stimulation, compared to neutrophils from non-infected mice (as displayed by both bioluminescence and FAM-FLICA fluorescence assays; Figs 9E and S7A , respectively). To directly assess the role of IFN-I nigericin-induced caspase1 activity, we inoculated mice with an IFN-I blocking anti-IFNAR monoclonal antibody or an isotype control antibody post-infection. Remarkably, neutrophils from anti-IFNAR treated mice showed diminished nigericin-induced caspase1 activity (Figs 9F and S7A ). Furthermore, as expected due to their typical IFN-responsiveness, oasl2, caspase1 and IL-1β gene expressions were lower in anti-IFNAR treated than in isotype treated mice ( Fig 9G–9I ). Taken together, the results indicate that IFN-I is responsible for the increased caspase1 activity in neutrophils of infected mice.
Blocking IFN-I signaling did not significantly alter virus replication, virus-induced pathological changes, neutrophil CD11b expression or neutrophil caspase1 activity without exogenous stimuli ( S7B–S7D , S7H and S7I Fig and S3 Table ). Neutrophil counts indicated a significant increase in neutrophil accumulation in the lungs of anti-IFNAR treated mice; however, this was not confirmed by neutrophil/lymphocyte ratio or morphometry (S7E-S7G Fig). Interestingly, regardless of treatment, some neutrophils in infected mice displayed degeneration and NETosis, evidenced by histone H3cit staining ( Fig 7J ; S3 Table ).
Neutrophils, the largest cell population of the host immune system, are rapidly recruited to sites of infection and play an important role in orchestrating an early immune response [ 42 , 43 ]. The relevance of neutrophils in viral infections became increasingly apparent during the COVID-19 pandemic, as they have been shown to be key mediators of the observed pathological processes [ 44 ].
This study sheds light on the potential involvement of the inflammasome pathway in COVID-19, particularly by demonstrating its activation in mature neutrophils during SARS-CoV-2 infection. Our investigation of the inflammatory profile of neutrophils as the dominant population of peripheral blood polymorphonuclear cells (PMNs) revealed an increased ability of neutrophils from severe COVID-19 patients for inflammasome assembly as evidenced by their transcriptional profile, spontaneous release of IL-1β, and elevated caspase1 activity. These findings are consistent with previous reports indicating activation of the NLRP3 inflammasome and ASC specks in circulating neutrophils during acute COVID-19 [ 14 , 16 ]. Furthermore, despite showing increased caspase1 activity, neutrophils from COVID-19 patients exhibited diminished soluble IL-1β production upon exogenous activation of the NLRP3 inflammasome pathway compared to healthy controls, which suggests that this pathway is “exhausted” due to prior activation during the disease. Mechanistically, our findings show that IFN-I, elevated in COVID-19 patients [ 45 , 46 ], can prime inflammasome formation in neutrophils. Transcriptomic analyses revealed that circulating neutrophils during severe COVID-19 show increased expression of IFN-responsive genes, suggesting inflammasome priming by IFN-I also in vivo during COVID-19 [ 47 ]. Furthermore, the study found that immature neutrophils, which are prevalent in low-density granulocyte fraction (LDGs), exhibit unique inflammasome gene expression and outcomes compared to mature neutrophils (PMNs). Distinctively from PMNs, LDGs do not display the IFN-I signature or upregulation of major inflammasome related genes, which indicates their lower responsiveness to IFN-I during COVID-19. However, since we were able to show that LDGs can form inflammasomes when stimulated by IFN-I ex vivo , their lower responsiveness is probably due to LDGs not being similarly exposed to IFN-I as compared to PMNs during COVID-19.
SARS-CoV-2 infected mice also showed increased neutrophil caspase1 activity, reversible by an IFN-I receptor (IFNAR) blocking antibody. Transcriptional analysis revealed a robust IFN-I signature and elevated expression of inflammasome genes encoding for caspase1 and IL-1β in neutrophils of infected mice, which were also inhibited by blocking IFNAR signaling, suggesting that IFN-I may also prime for inflammasome activation in mice. Notably, the anti-IFNAR treatment did not affect neutrophil recruitment or NETosis, which is consistent with another COVID-19 model using transgenic human ACE2, where IFNAR knockout inhibited recruitment of monocytes and lymphocytes, but not neutrophils, to infected lungs [ 48 ].
Inflammasomes were first studied in macrophages, revealing many molecular mechanisms regulating inflammasome assembly [ 49 ]. Macrophage inflammasome activation has emerged as a major factor also in COVID-19 [ 17 ]. Interestingly, macrophage inflammasome activation was recognized to be IFN-I mediated in an experimental rhesus macaque COVID-19 model [ 50 ]. However, due to the abundance of neutrophils compared with cells of monocyte/macrophage lineage [ 51 , 52 ], the significance of neutrophil inflammasomes in COVID-19 is likely underestimated. Our results highlight inflammasomes as an additional important inflammatory mechanism in neutrophils [ 14 ], complementing their role in phagocytosis, reactive oxygen species generation, degranulation, and NETosis [ 31 ].
SARS-CoV-2 can directly activate inflammasomes in cells of the monocyte/macrophage lineage [ 17 ]. Our study investigated whether SARS-CoV-2 can provide the first or second signal for inflammasome activation in neutrophils. However, we found no evidence of direct virus-induced inflammasome activation in neutrophils. The difference between macrophages and neutrophils in their susceptibility to SARS-CoV-2 could depend on many factors. Both cell types express ACE2, the receptor for SARS-CoV-2, but may differ in ACE2 expression levels [ 53 ]. Furthermore, the intracellular environment of macrophages is better suited for viral replication [ 54 ], while neutrophils focus on phagocytosis and antimicrobial responses [ 31 , 55 ]. Additionally, pathogen opsonization can trigger inflammasomes in macrophages [ 56 ] but is not a primary function of neutrophils. Therefore, our findings suggest neutrophil inflammasome activation in response to SARS-CoV-2 likely results from interactions with infected and/or dying cells in the lungs, rather than direct virus activation. To note, whether SARS-CoV-2 can induce neutrophil inflammasomes through immune complex-mediated mechanisms, as seen in monocytes/macrophages [ 17 ] remains to be determined.
In this study, we demonstrated IFN-I as the first signal for NLRP3 inflammasome activation in neutrophils. While prior research has explored IFN-inflammasome crosstalk [ 57 ], priming capacity of IFN-I remained unclear. While IFN-I promotes inflammasomes in epithelial cells [ 58 ] it can also dampen IL-1β in macrophages [ 59 ]. Plausibly, initial IFN-I exposure may upregulate inflammasome genes, whereas prolonged activity could hinder IFN-I signaling via “negative feedback” loop, in line with our findings of inflammasome exhaustion in circulating neutrophils of severe COVID-19 patients. It should be noted that several SARS-CoV-2 encoded proteins have been shown to inhibit IFN-I signaling [ 60 ]. However, no evidence suggests that neutrophils can be infected by SARS-CoV-2 and therefore it seems unlikely that such direct virus mediated effects could play a role in the observed neutrophil unresponsiveness to IFN-I.
The dualistic nature of the IFN-I response in COVID-19 has been recognized previously. It seems that a strong initial IFN-I response to SARS-CoV-2 is more likely to result in asymptomatic or mild COVID-19 whereas a decreased initial IFN-I activity, due to e.g. genetic defects or increased levels of IFN-I autoantibodies, can lead to more severe COVID-19 [ 61 ]. This initial beneficial effect of IFN-I is probably due to its ability to limit viral replication at early stages of the infection. However, at later stages of the disease IFN-I can be detrimental by promoting inflammatory pathways instead of direct antiviral effects [ 62 ]. Thus, similarly to the IFN-I response in general, the role of neutrophil inflammasomes in development and severity of COVID-19 might be dualistic in nature with an initial protective effect while damaging when sustained for prolonged periods.
Our study demonstrated a strong association between PMN caspase1 activity and plasma levels of calprotectin, a marker of neutrophil activation. It is of interest to note that calprotectin can also promote inflammasome activity in neutrophils [ 63 , 64 ]. Therefore, in addition to IFN-I discussed in this study, it is possible that calprotectin also contributes to neutrophil inflammasome formation during COVID-19. Additionally, increased disease severity, as assessed by the WHO ordinal scale, was significantly linked to PMNs being less responsive to ex vivo IFN-induced inflammasome activation. Thus, these results suggest that neutrophil inflammasomes would play a role in disease severity, rather than being protective in COVID-19.
Our study also unveiled distinct gene profiles in LDGs and PMNs from severe COVID-19 patients. LDGs exhibited upregulation of genes related to DNA replication and cell cycle, indicating immaturity, and confirming our prior findings [ 7 ]. Conversely, PMNs displayed heightened NLR signaling, suggesting a robust response to pathogens. While our study compared PMNs and LDGs, and the COVID-19 Immune Atlas single cell analysis represented a broader classification of mature and immature neutrophils, the alignment of our results with the atlas provides further support for the distinct characteristics of these two neutrophil populations in severe COVID-19. Notably, IL-18 gene expression and secretion after ex vivo stimulation were higher in LDGs than PMNs. To note, PMN’s lack of IL-18 secretion is not due to lack of protein, as they constitutively express significant amounts intracellularly [ 65 ]. This indicates a similarity between LDGs and monocytes/macrophages in inflammasome mediated IL-18 processing, possibly lost during neutrophil maturation.
The present study has some limitations worth discussing. Firstly, the relatively small human sample size may limit the generalizability of the findings. While RNA-seq provided valuable insights into gene expression profiles of PMNs and LDGs, we did not perform functional validation of the identified pathways in this study. Regarding our experimental SARS-CoV-2 disease model, the high virus input might trigger robust immune responses that differ from typical human infections, and the short-lived virus replication in the applied model does not capture the effect of prolonged antigen exposure or the complex inflammatory milieu seen in human cases. Importantly, our results do not directly assess the role of neutrophil inflammasomes in COVID-19 pathogenesis in humans or in the animal disease model. Further studies are therefore needed to understand the relative contribution of neutrophil inflammasomes in COVID-19 disease progression, compared to the better described macrophage inflammasomes as well as to other inflammatory pathways engaged by neutrophils such as degranulation, reactive oxygen species production and NETosis. Furthermore, due to the ubiquitous expression of IFNAR, the observed inhibitory effects on neutrophil inflammasome activity by IFNAR blockade does not exclude the possibility that IFN-I could promote neutrophil inflammasome formation by indirect effects such as regulating the interplay between neutrophils and other immune cells or stimulating the release of pro-inflammatory cytokines by other cell types. In addition, investigating the effects of IFNAR blockade in other time points than the chosen 2 dpi might have been more valuable in revealing its effects on viral replication and neutrophil accumulation in the infected lungs. Finally, since the prominent role of neutrophils in the immune response to viral infections is widely recognized [ 42 , 43 ] and it would be valuable to compare these findings to neutrophil responses in other viral respiratory infections.
Taken together, our findings provide valuable insights into neutrophil involvement in COVID-19 and possibly other viral respiratory infections. However, further research is needed to fully grasp the role of neutrophil inflammasomes in COVID-19 pathogenesis. This increased understanding may facilitate the development of targeted treatment approaches for COVID-19. For example, pharmacologically targeting the inflammasome pathway in neutrophils with novel inhibiting molecules [ 66 ], may help mitigate the exaggerated inflammatory response observed in severe cases. The next steps involve validating the pathways and genes identified as potential therapeutic targets and assessing their COVID-19 specificity. Prospectively, these strategies could be extended to address upcoming respiratory virus pandemics, where neutrophils and inflammasomes provide major pathogenic contributions.
Supporting information
S1 fig. comparison of gene expression in granulocyte populations of covid-19 patients using rna-seq analysis..
( A ) Deconvoluted RNA-seq data. The cellular composition in isolated PMN and LDG fractions was estimated using CIBERSORTx through the identification of cell populations based on RNA-seq. The bar plots in the figure represent the cell composition of each RNA-seq sample, offering insights on sample purity. ( B ) Heatmap of the top 118 differentially expressed genes between PMNs from healthy controls, mild and severe COVID-19, as well as LDGs from severe disease, identified by unsupervised ICGS analysis based on correlation, using AltAnalyze software. IFN-related genes, identified by GENESHOT, are shown in bold.
https://doi.org/10.1371/journal.ppat.1012368.s001
S2 Fig. Enriched differentially expressed genes and pathways in severe COVID-19 PMNs and LDGs.
( A-B ) Volcano plots of DEGs between severe COVID-19 PMNs versus ( A ) severe COVID-19 LDGs and ( B ) HC PMNs. ( C-D ) Volcano plots of enriched gene sets in severe COVID-19 PMNs versus ( C ) HC PMNs and ( D ) severe COVID-19 LDGs, using KEGG database. Each point represents a single gene set, where the x-axis measures its odds ratio, while the y-axis shows its -log10(p-value). ( E-F ) Volcano plots of ( E ) severe COVID-19 PMNs versus mild COVID-19 PMNs and ( F ) mild COVID-19 PMNs vs HC PMN. For all panels, blue points represent significant terms (adjusted p-value < 0.05), while smaller gray points represent non-significant terms. DEG = differentially expressed genes .
https://doi.org/10.1371/journal.ppat.1012368.s002
S3 Fig. Differential expression of interferon and inflammasome related genes in PMNs during COVID-19.
RNA was extracted from isolated HC PMNs (n = 8–13) versus severe COVID-19 PMNs (n = 29–32) and subjected to comparative RT-qPCR using specific primers for OAS1, OAS2, IFIT1, IFI16, caspase1, caspase5, IL1B, NLRC4, NLRC5, NLRP3 and NAIP. *p < 0.05, **p < 0.01, ***p < 0.001 and **** p < 0.0001. P values calculated with Mann-Whitney U-test. Data presented as mean ± SD.
https://doi.org/10.1371/journal.ppat.1012368.s003
S4 Fig. Expression of inflammasome related genes in mature and immature neutrophils from COVID-19 PBMCs.
The fraction of mature and immature neutrophils cells expressing 17 inflammasome related genes identified in Fig 2D (shown in black and blue, respectively) are shown in a bar graph. For each gene, the proportion of expressing cells is shown in light blue, while the proportion of negative or not-expressing cells is shown in gray. Zoomed-in bar graph depicts the proportion of mature and immature cells expressing each gene.
https://doi.org/10.1371/journal.ppat.1012368.s004
S5 Fig. Ex vivo stimulation of isolated PMNs.
( A ) HC PMNs (1 million/ml) were primed for 4 hr by low dose IFN-α, low dose IFN-β (both 2.7*10 3 IU/ml) or 20 ng/ml LPS followed by 2.5 μM nigericin activation for 4 hr. IL-1β release was measured by ELISA and the assays were performed in the presence of either α-IFNAR1 or mouse IgG as control (both 100 μg/ml) (n = 5). *p < 0.05 and **p < 0.01. P values calculated using two-way ANOVA with Šídák multiple comparison test. ( B ) IL-18 (n = 2–3 HC PMN and 3 COVID-19 PMN) was measured from supernatants by ELISA following LPS or IFN-I priming (4 h) and subsequent nigericin activation (4 h). ( C-E ) Effect of different inflammasome specific inhibitors in cytokine secretion. ( C ) Effect of inflammasome inhibitor MCC950 (2 μg/ml) and YVAD (20 μg/ml) on LPS or IFN-I primed (4 h) and nigericin activated (4 h) IL-1β secretion in the supernatant of healthy control PMNs (n = 8). ( D-E ) Effect of inflammasome inhibitor MCC950 (2 μg/ml, added simultaneously with nigericin) on LPS or IFN-I primed (4 h) and nigericin activated (20 h). *p < 0.05, **p < 0.01, ***p < 0.001, **** p < 0.0001. P values calculated with Kruskal-Wallis test. Data presented as mean ± SD. IFN = interferon type I , LPS = lipopolysaccharide , Nig = nigericin , YVAD = tetrapeptide caspase1 inhibitor Tyr-Val-Ala-Asp . ( F-I ) Gene expressions in HC and COVID-19 PMNs after LPS or IFN-I stimulation. A comparison of gene expression in isolated healthy control PMNs versus COVID-19 PMNs after ex vivo stimulation with LPS or IFN-I. Extracted RNA was subjected to comparative RT-qPCR using specific primers for NLRP3, NLRC4, NAIP and CASP5 (n = 4–8 for HC PMN and 6–9 for COVID-19 PMN). *p < 0.05. Two-way ANOVA with Tukey’s multiple comparison test was applied. Data were presented as mean ± SD.
https://doi.org/10.1371/journal.ppat.1012368.s005
S6 Fig. Animal weight dynamics, flow cytometry gating strategy and immunohistochemistry in SARS-CoV-2 infected mice.
Female BALB/c mice were intranasally inoculated with 5*10 5 TCID50 SARS-CoV-2 MaVie strain or PBS as control and euthanized at 2 dpi or 4 dpi. ( A ) Daily tracking of animal weight performed throughout the experiment (n = 12 for SARS-CoV-2 infected animals, n = 6 for PBS-inoculated animals). The weights of the mice euthanized at 2 dpi (n = 26) did not show significant differences and are not reported. ( B ) Gating strategy to analyze Ly6G+ neutrophils in mouse lung single cell suspensions. Side scatter area (SSC-A) versus forward scatter area (FSC-A) plot followed by side scatter area versus height (SSC-A vs SSC-H) plot were used for the identification of single cells. BV605 yellow live/dead dye was used to discriminate dead cells, from which CD3/CD19+ lymphocytes and Ly6G+ neutrophils were gated as shown. ( C ) Representative histogram showing the percentage of Ly6G+ cells after isolation from lung single cell suspension using Ly6G-binding magnetic beads. ( D ) Left column: immunohistochemistry for SARS-CoV-2 nucleoprotein; right column: immunohistochemistry for Ly6G (neutrophil marker), hematoxylin counterstain. Bars = 500 μm (large images) and 50 μm (insets). At 2 dpi (top), the arrow points at a bronchus with viral antigen expression in epithelial cells. A close–up of the bronchus (bottom; B: bronchial lumen) shows degenerated and slough off antigen positive epithelial cells. Adjacent alveoli exhibit viral antigen expression in typeI (arrowhead) and typeII (arrow) pneumocytes. The overview (top) shows neutrophils between the infected bronchial (arrow) epithelial cells, in parenchymal areas (arrowhead; right inset) and in capillaries (arrowheads). A close-up of the bronchus (bottom; B: bronchial lumen) highlights numerous neutrophils between degenerate (arrowheads) epithelial cells. At 4 dpi (middle), there are focal areas with antigen expression in alveolar epithelial cells and infiltrating macrophages. Neutrophils are present among the infiltrating cells (arrow) as individual cells (inset: arrows) or in aggregates (inset: arrowhead). The bottom shows the lung of a mock-infected control animal. There is no viral antigen expression. Staining for Ly6G depicts individual neutrophils in larger vessels (inset: arrow) or in capillaries (inset: arrowheads).
https://doi.org/10.1371/journal.ppat.1012368.s006
S7 Fig. Impact of α-IFNAR treatment in SARS-CoV-2 infected mice.
Female BALB/c mice were intranasally inoculated with 5* 10 5 TCID50 SARS-CoV-2 MaVie strain or PBS as control. Mice were intraperitoneally inoculated with 250 μg anti-IFNAR or IgG1 isotype control directly after infection with SARS-CoV-2 and lung neutrophils isolated at 2 dpi (including also intranasally PBS-inoculated control mice without intraperitoneal injection) ( A ) Quantification of caspase1 positive cells in nigericin-activated isolated Ly6G neutrophils stained by FAM-FLICA. Representative histogram is shown. * p < 0.05. P values calculated using one-way ANOVA with multiple comparison test (Holm-Šídák correction). ( B ) RNA was isolated from mouse lungs and subjected to RT-qPCR targeting the replication-intermediate subgenomic E gene and GAPDH as housekeeping gene. RNA levels were assessed based on cycle threshold Ct levels. The expression levels of the target gene SubE were measured and normalized to GAPDH levels using the comparative Ct method (ΔΔCt). The fold change values were calculated by the formula 2^(-ΔΔCt), representing the relative gene expression compared to the PBS mock-infected control (in which subE was undetectable but set to 40 Ct). No significant differences are seen between the two groups, assessed with Welch’s t-test. ( C ) Infectious virus was calculated from supernatants of lung single cell suspensions of infected mice as fluorescence focus forming units (FFU) in Vero E6 cells. ( D ) Quantification based on morphometric analysis that determines the area of immunolabelling for SARS-CoV-2 nucleoprotein in relation to total tissue area. ( E ) Quantification of Ly6G neutrophil/lymphocyte ratio in lung single cell suspensions by flow cytometry. ( F ) Quantification of Ly6G cell counts extrapolated per lung in single cell suspensions by flow cytometry ( G ) Quantification of Ly-6G based on morphometric analysis that determines the area of immunolabelling for Ly6G in relation to total tissue area in mock-infected controls. ( H ) Quantification of median fluorescence intensity (MFI) of CD11b expression in ly6G neutrophils by flow cytometry. ( I ) Quantification of FAM-FLICA MFI in Ly6G+ neutrophils by flow cytometry. Panels B, D and G are representative of two independent experiments. ( J ) Histological features, viral antigen expression and extent of neutrophil influx and evidence of neutrophil damage in the lung of SARS-CoV-2 infected BALB/C mice after isotype control and anti-IFNAR treatment at 2dpi. Left column: Control isotype treated mice; right column: anti-IFNAR treated mice. HE stain (top layer) and immunohistology, hematoxylin counterstain (all other images). Bars: 250 μm (overview images) and 25 μm (insets). In control isotype treated mice, the lung exhibits degeneration and loss of bronchial and bronchiolar epithelial cells (HE stain: arrowhead; right inset), with mild inflammatory infiltration. The parenchyma exhibits focal areas of increased cellularity, with typeII pneumocyte activation and occasional degenerate alveolar epithelial cells (arrows; left inset: degenerate cells (arrowhead) and infiltrating neutrophil (arrow)). Staining for SARS-CoV-2 NP confirms epithelial cell infection in bronchus (arrowhead; right inset) and alveoli (arrow; left inset). Right inset: Viral antigen expression is seen in intact and sloughed off, degenerate epithelial cells. Left inset: Viral antigen expression is seen in both typeI (small arrowhead) and typeII (small arrow) pneumocytes; there are also degenerate positive cells (large arrowhead). Neutrophils (Ly6G+) are located within focal parenchymal areas of increased cellularity (arrows; left inset: arrowheads) and present between degenerate bronchial epithelial cells (arrowhead; right inset: arrowhead). Staining for histone H3 shows neutrophil degeneration/NETosis in parenchymal areas (arrow; left inset: arrowheads) and associated with degenerate epithelial cells (arrowhead; right inset: positive reaction between sloughed off epithelial cells (arrow) and between the intact epithelial layer (arrowhead)). In anti-IFNAR treated animals, the lung exhibits degeneration and loss of bronchial and bronchiolar epithelial cells (arrowhead; right inset: arrows), with mild inflammatory infiltration and individual neutrophils between intact and sloughed off degenerate epithelial cells (right inset: arrowheads). The parenchyma exhibits focal areas of increased cellularity, with typeII pneumocyte activation and occasional degenerate alveolar epithelial cells (arrows; left inset: degenerate cells (arrow) and infiltrating neutrophils (arrowhead). Staining for SARS-CoV-2 NP shows epithelial cell infection in bronchioles (arrowhead; right inset) and alveoli (arrow; left inset). Right inset: Viral antigen expression is seen in intact and sloughed off, degenerate epithelial cells. Left inset: Viral antigen expression is seen in pneumocytes (arrow) and infiltrating macrophages (arrowheads). Neutrophils (Ly6G+) locate within focal parenchymal areas of increased cellularity (arrows; left inset) and are present between intact (inset: arrowhead) and degenerate epithelial cells (arrowhead; right inset: arrow). Staining for histone H3cit shows neutrophil degeneration/NETosis in parenchymal areas (arrow; left inset) and associated with degenerate epithelial cells (arrowhead; right inset: positive reaction between sloughed off epithelial cells (arrow) and between the intact epithelial layer (arrowhead)). Dpi = days post infection; NP = nucleoprotein .
https://doi.org/10.1371/journal.ppat.1012368.s007
S1 Table. Information for Fig 3 , detailing the Gene Set Enrichment Analysis (GSEA) databases used for pathway analyses.
https://doi.org/10.1371/journal.ppat.1012368.s008
S2 Table. qPCR primer sequences: gene-specific forward and reverse primers.
https://doi.org/10.1371/journal.ppat.1012368.s009
S3 Table. Histological changes as well as SARS-CoV-2 nucleoprotein and RNA expression in female BALB/C mice infected with SARS-CoV-2.
https://doi.org/10.1371/journal.ppat.1012368.s010
S1 Source Data. Original scan of IL-1β and actin western blot. HC PMNs were non-stimulated or stimulated 4h with IFN-I (combination of 2.7*104 IU/ml IFN-α and IFN-β) or 20 ng/ml LPS (1st signal), followed by 4h with 2.5 μM nigericin (2nd signal).
Western blots from supernatants and cell lysates were performed first for actin and then for IL-1β on the same membrane as indicated. Information for Fig 5A .
https://doi.org/10.1371/journal.ppat.1012368.s011
Acknowledgments
RNA isolation, library preparations and RNA sequencing was performed at the Institute for Molecular Medicine Finland FIMM, Genomics unit supported by HiLIFE and Biocenter Finland. The authors also thank M. Utriainen for expert technical assistance.
- View Article
- PubMed/NCBI
- Google Scholar
- 25. CZ CELLxGENE Discover. Chan Zuckerberg Initiative. Available: https://cellxgene.cziscience.com/

Aug. 22, 2024
Is there support for a houston independent school district bond, brief : aug. 22, 2024 education.

This study seeks to understand public support for the Houston Independent School District's proposed $4.4 billion bond to address infrastructure and education needs.
The Houston Independent School District (HISD) is seeking a $4.4 billion bond aimed at addressing critical infrastructure and educational needs. Branded as “ Renew HISD ,” the package would rebuild and modernize over 40 campuses, upgrade HVAC systems, improve campus security and expand early childhood and career and technical education programs.
As the district was preparing the proposal, the Kinder Institute for Urban Research conducted a survey in January to understand public support for education bonds. Results of that survey were shared with district officials, and a follow-up survey was conducted in August to determine whether public support had changed. This brief summarizes the main findings of the January survey with additional discussion of what was learned in the August follow-up survey.
Key findings
- Support for education bonds is high among residents living in HISD, but only if it increases property taxes marginally. Support did not substantially increase between January and August.
- HISD residents overall identified updated campus safety and security measures and additional career and technical education facilities as priorities.
- HISD residents expressed a strong interest in being directly involved in decision making.
- Most respondents agreed that Houston-area public schools need significantly more funding overall to provide a quality education.
Voters living in HISD will decide on the proposed bond on the November ballot.

Mailing Address
6100 Main St. MS-208 Houston, TX 77005-1892
[email protected] 713-348-4132
Subscribe to our e-newsletter
Physical Address
Rice University Kraft Hall 6100 Main Street, Suite 305 Houston, TX 77005-1892
Featured Sponsor
Support the Kinder Institute

IMAGES
COMMENTS
Research results refer to the findings and conclusions derived from a systematic investigation or study conducted to answer a specific question or hypothesis. These results are typically presented in a written report or paper and can include various forms of data such as numerical data, qualitative data, statistics, charts, graphs, and visual aids.
The Results section of a scientific research paper represents the core findings of a study derived from the methods applied to gather and analyze information. It presents these findings in a logical sequence without bias or interpretation from the author, setting up the reader for later interpretation and evaluation in the Discussion section.
A results section is where you report the main findings of the data collection and analysis you conducted for your thesis or dissertation. You should report all relevant results concisely and objectively, in a logical order. ... Reporting qualitative research results. In qualitative research, your results might not all be directly related to ...
Research findings refer to the results obtained from a study or investigation conducted through a systematic and scientific approach. These findings are the outcomes of the data analysis, interpretation, and evaluation carried out during the research process.
The results section should state the findings of the research arranged in a logical sequence without bias or interpretation. A section describing results should be particularly detailed if your paper includes data generated from your own research. ... It is appropriate to highlight this finding in the results section. However, speculating as to ...
The results section of a quantitative research paper is where you summarize your data and report the findings of any relevant statistical analyses. The APA manual provides rigorous guidelines for what to report in quantitative research papers in the fields of psychology, education, and other social sciences.
The results chapter (also referred to as the findings or analysis chapter) is one of the most important chapters of your dissertation or thesis because it shows the reader what you've found in terms of the quantitative data you've collected. It presents the data using a clear text narrative, supported by tables, graphs and charts.
Present the results of the paper, in logical order, using tables and graphs as necessary. Explain the results and show how they help to answer the research questions posed in the Introduction. Evidence does not explain itself; the results must be presented and then explained. Avoid: presenting results that are never discussed; presenting ...
1. Context. The "results section" is the heart of the paper, around which the other sections are organized ().Research is about results and the reader comes to the paper to discover the results ().In this section, authors contribute to the development of scientific literature by providing novel, hitherto unknown knowledge ().In addition to the results, this section contains data and ...
Developing a well-written research paper is an important step in completing a scientific study. This paper is where the principle investigator and co-authors report the purpose, methods, findings, and conclusions of the study. A key element of writing a research paper is to clearly and objectively report the study's findings in the Results section.
The Results (also sometimes called Findings) section in an empirical research paper describes what the researcher(s) found when they analyzed their data. Its primary purpose is to use the data collected to answer the research question(s) posed in the introduction, even if the findings challenge the hypothesis.
The dissertation's findings section presents the key results of your research without interpreting their meaning. Theoretically, this is an exciting section of a dissertation because it involves writing what you have observed and found. However, it can be a little tricky if there is too much information to confuse the readers.
The "Results" section is arguably the most important section in a research manuscript as the findings of a study, obtained diligently and painstakingly, are presented in this section. A well-written results section reflects a well-conducted study. This chapter provides helpful pointers for writing an effective, organized results section.
The results chapter in a dissertation or thesis (or any formal academic research piece) is where you objectively and neutrally present the findings of your qualitative analysis (or analyses if you used multiple qualitative analysis methods ). This chapter can sometimes be combined with the discussion chapter (where you interpret the data and ...
The researcher usually organizes the results of his/her results section by research question or hypothesis, stating the results for each one, using statistics to show how the research question or hypothesis was answered in the study. ... and explains what the findings mean. If the results confirmed or corresponded with the findings of other ...
The results section of a research paper tells the reader what you found, while the discussion section tells the reader what your findings mean. The results section should present the facts in an academic and unbiased manner, avoiding any attempt at analyzing or interpreting the data. Think of the results section as setting the stage for the ...
The "Results section" should focus on the findings that answer the research question(s), regardless of whether the findings were positive or negative, and whether they are statistically significant or not. 8,11 Whenever possible, it is best to present the main findings using figures as readers are mostly attracted by the figures and ...
Technically or academically speaking, 'findings' seems to be used more for qualitative studies whereas 'results' seems to be used more for quantitative studies. For example, in a behavioral study, you may say 'It was found that when service executives aimed to build rapport with customers, it led to purchases 72% of the time compared ...
The results section of the research paper is where you report the findings of your study based upon the information gathered as a result of the methodology [or methodologies] you applied. The results section should simply state the findings, without bias or interpretation, and arranged in a logical sequence. The results section should always be ...
Laura Moro-Martin, freelance scientific writer on Kolabtree, provides expert tips on how to write the results section of a research paper. You have prepared a detailed −but concise− Methods section.Now it is time to write the Results of your research article. This part of the paper reports the findings of the experiments that you conducted to answer the research question(s).
3). Research Questions as Headings . You can also present your findings using your research questions as the headings in the findings section. This is a useful strategy that ensures you're answering your research questions and also allows the reader to quickly ascertain where the answers to your research questions are.
The discussion section provides an analysis and interpretation of the findings, compares them with previous studies, identifies limitations, and suggests future directions for research. This section combines information from the preceding parts of your paper into a coherent story. By this point, the reader already knows why you did your study ...
The interpretation of results in research requires multiple steps, including checking, cleaning, and editing data to ensure its accuracy, and properly organizing it in order to simplify interpretation. To examine data and derive reliable findings, researchers must employ suitable statistical methods.
The findings prompt further investigation into whether there exist distinct subgroups of adolescents who exhibit similar patterns of anxiety and depression under stress. Future research could extend this inquiry to better understand the nuanced relationships between stress, anxiety, and depression among adolescents. Limitations
The results show that macrophage ILF3 deficiency attenuates abdominal aortic aneurysm progression, while elevated macrophage ILF3 exacerbates abdominal aortic aneurysm lesions. ... our findings ...
Introduction: Prader-Willi Syndrome (PWS), a rare genetic disorder, affects development and behavior, frequently resulting in self-injury, aggression, hyperphagia, oppositional behavior, impulsivity and over-activity causing significant morbidity. Currently, limited therapeutic options are available to manage these neuropsychiatric manifestations. The aim of this clinical trial was to assess ...
Study findings may help explain why we keep getting colds By simultaneously circulating many versions of itself, the rhinovirus ups the odds that it will latch onto people who are not immune. August 21, 2024. ... a postdoctoral research scientist in the Greninger lab. She was lead author of the paper about the findings.
Purdue University
Author summary COVID-19, caused by the SARS-CoV-2, ranges from mild "flu"-like symptoms to severe respiratory distress or even death. Neutrophils are important cells of our immune system which are strongly involved in inflammatory responses, including those occurring in COVID-19. However, despite extensive research, the precise contribution of neutrophils to the pathogenesis of COVID-19 ...
Results of that survey were shared with district officials, and a follow-up survey was conducted in August to determine whether public support had changed. This brief summarizes the main findings of the January survey with additional discussion of what was learned in the August follow-up survey. Key findings