An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
- My Bibliography
- Collections
- Citation manager

Save citation to file
Email citation, add to collections.
- Create a new collection
- Add to an existing collection
Add to My Bibliography
Your saved search, create a file for external citation management software, your rss feed.
- Search in PubMed
- Search in NLM Catalog
- Add to Search
A case study of multiple-treatments meta-analysis demonstrates that covariates should be considered
Affiliation.
- 1 MRC Biostatistics Unit, Cambridge, UK. [email protected]
- PMID: 19157778
- DOI: 10.1016/j.jclinepi.2008.10.001
Objective: To illustrate the potential and challenges of the simultaneous analysis of a network of trials, using as a case study the investigation of the relative effectiveness of four topical fluoride treatments and two control interventions (placebo and no treatment) in preventing dental caries in children.
Study design and setting: We performed multiple-treatments meta-analysis within a Bayesian framework by synthesizing six Cochrane reviews. We explored the compatibility between direct and indirect evidence and adjusted the results using a meta-regression model to take into account differences in the year of randomization across studies.
Results: The validity of our conclusions for the superiority of fluoride toothpaste as indicated from the initial network analysis using Bayesian methods was challenged when we adjusted for possible confounders. The network was dominated by studies comparing placebo with toothpaste, which were older and had been carried out in populations with higher baseline risk than studies involving other fluoride modalities.
Conclusion: After adjusting for possible differences across studies, we did not find clear evidence that any topical fluoride modality is more effective than any other. Multiple-treatments meta-analysis methods allow for more detailed investigations than naïve methods in the analysis of indirect evidence on treatment effects.
PubMed Disclaimer
Similar articles
- Fluoride toothpastes of different concentrations for preventing dental caries in children and adolescents. Walsh T, Worthington HV, Glenny AM, Appelbe P, Marinho VC, Shi X. Walsh T, et al. Cochrane Database Syst Rev. 2010 Jan 20;(1):CD007868. doi: 10.1002/14651858.CD007868.pub2. Cochrane Database Syst Rev. 2010. Update in: Cochrane Database Syst Rev. 2019 Mar 04;3:CD007868. doi: 10.1002/14651858.CD007868.pub3. PMID: 20091655 Updated. Review.
- Cochrane reviews of randomized trials of fluoride therapies for preventing dental caries. Marinho VC. Marinho VC. Eur Arch Paediatr Dent. 2009 Sep;10(3):183-91. doi: 10.1007/BF03262681. Eur Arch Paediatr Dent. 2009. PMID: 19772849 Review.
- Fluoride toothpastes of different concentrations for preventing dental caries. Walsh T, Worthington HV, Glenny AM, Marinho VC, Jeroncic A. Walsh T, et al. Cochrane Database Syst Rev. 2019 Mar 4;3(3):CD007868. doi: 10.1002/14651858.CD007868.pub3. Cochrane Database Syst Rev. 2019. PMID: 30829399 Free PMC article.
- Combinations of topical fluoride (toothpastes, mouthrinses, gels, varnishes) versus single topical fluoride for preventing dental caries in children and adolescents. Marinho VC, Higgins JP, Sheiham A, Logan S. Marinho VC, et al. Cochrane Database Syst Rev. 2004;2004(1):CD002781. doi: 10.1002/14651858.CD002781.pub2. Cochrane Database Syst Rev. 2004. PMID: 14973992 Free PMC article. Review.
- Cochrane reviews on the benefits/risks of fluoride toothpastes. Wong MC, Clarkson J, Glenny AM, Lo EC, Marinho VC, Tsang BW, Walsh T, Worthington HV. Wong MC, et al. J Dent Res. 2011 May;90(5):573-9. doi: 10.1177/0022034510393346. Epub 2011 Jan 19. J Dent Res. 2011. PMID: 21248357 Review.
- Efficacy of Local Anesthesia for Radial Artery Puncture Pain: A Systematic Review and Network Meta-Analysis. Yasuo S, Hayashi M, Suda C, Kataoka Y, Taito S, Imai E, Sazanami K. Yasuo S, et al. Cureus. 2024 Jul 16;16(7):e64682. doi: 10.7759/cureus.64682. eCollection 2024 Jul. Cureus. 2024. PMID: 39149654 Free PMC article. Review.
- Effectiveness of Rehabilitation for Osteoarthritis of the Knee: A Scoping Review of Network Meta-Analyses. Kitagawa T, Denda T, Okuyama W, Miyachi R, Nakamura K. Kitagawa T, et al. Cureus. 2024 Apr 5;16(4):e57661. doi: 10.7759/cureus.57661. eCollection 2024 Apr. Cureus. 2024. PMID: 38707059 Free PMC article. Review.
- Nutritional Interventions for the Prevention of Cognitive Decline in Patients With Mild Cognitive Impairment and Alzheimer Disease: Protocol for a Network Meta-Analysis of Randomized Controlled Trials. He Q, Wu KCH, Bennett AN, Zhang JY, Chan KHK. He Q, et al. JMIR Res Protoc. 2024 Feb 28;13:e47196. doi: 10.2196/47196. JMIR Res Protoc. 2024. PMID: 38416536 Free PMC article.
- Methodological review of NMA bias concepts provides groundwork for the development of a list of concepts for potential inclusion in a new risk of bias tool for network meta-analysis (RoB NMA Tool). Lunny C, Veroniki AA, Higgins JPT, Dias S, Hutton B, Wright JM, White IR, Whiting P, Tricco AC. Lunny C, et al. Syst Rev. 2024 Jan 12;13(1):25. doi: 10.1186/s13643-023-02388-x. Syst Rev. 2024. PMID: 38217041 Free PMC article.
- Systemic corticosteroids for the prevention of bronchopulmonary dysplasia, a network meta-analysis. Hay S, Ovelman C, Zupancic JA, Doyle LW, Onland W, Konstantinidis M, Shah PS, Soll R. Hay S, et al. Cochrane Database Syst Rev. 2023 Aug 31;8(8):CD013730. doi: 10.1002/14651858.CD013730.pub2. Cochrane Database Syst Rev. 2023. PMID: 37650547 Review.
- Search in MeSH
Related information
- Cited in Books
- PubChem Compound (MeSH Keyword)
Grants and funding
- MC_U105285807/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full text sources.
- Elsevier Science
- MedlinePlus Health Information
- Citation Manager
NCBI Literature Resources
MeSH PMC Bookshelf Disclaimer
The PubMed wordmark and PubMed logo are registered trademarks of the U.S. Department of Health and Human Services (HHS). Unauthorized use of these marks is strictly prohibited.
Jump to navigation

Cochrane Methods Methodology Register
| Author(s) | |
| Title | |
| Abstract | |
| Source | |
| CMR keyword(s) | |
| Earliest year | Latest year |
The Cochrane Methodology Register (CMR) is a bibliography of publications that report on methods used in the conduct of controlled trials. It includes journal articles, books, and conference proceedings, and the content is sourced from MEDLINE and hand searches. CMR contains studies of methods used in reviews and more general methodological studies that could be relevant to anyone preparing systematic reviews. CMR records contain the title of the article, information on where it was published (bibliographic details), and, in some cases, a summary of the article. They do not contain the full text of the article.
The CMR was produced by the Cochrane UK , until 31 st May 2012. There are currently no plans to reinstate the CMR and it is not receiving updates.* If you have any queries, please contact the Cochrane Community Service Team ( [email protected] ).
The Publishers, John Wiley & Sons Ltd, thanks Update Software for the continued use of their data formats in the Cochrane Methodology Register (CMR).
*Last update in January 2019.
| Title | |
| Authors | |
| Source | |
| Date of publication | |
| Volume | |
| Issue | |
| Pages | |
| Abstract | |
| CMR keywords | |
| Correspondence address | |
| Reference type | Journal article |
Europe PMC requires Javascript to function effectively.
Either your web browser doesn't support Javascript or it is currently turned off. In the latter case, please turn on Javascript support in your web browser and reload this page.
A case study of multiple-treatments meta-analysis demonstrates that covariates should be considered.
Full text links.
Add to Saved Papers
Get 1-tap access
Related Resources
For the best experience, use the read mobile app.
Get seemless 1-tap access through your institution/university For the best experience, use the Read mobile app
- DOI: 10.1016/j.jclinepi.2008.10.001
- Corpus ID: 8052945
A case study of multiple-treatments meta-analysis demonstrates that covariates should be considered.
- G. Salanti , V. Marinho , J. Higgins
- Published in Journal of Clinical… 1 August 2009
Figures and Tables from this paper

282 Citations
Incorporating multiple interventions in meta-analysis: an evaluation of the mixed treatment comparison with the adjusted indirect comparison, estimation and adjustment of bias in randomized evidence by using mixed treatment comparison meta‐analysis.
- Highly Influenced
Importance of assessing and adjusting for cross-study heterogeneity in network meta-analysis: a case study of psoriasis.
Meta-regression models to address heterogeneity and inconsistency in network meta-analysis of survival outcomes, incorporating adjustments for variability in control group response rates in network meta-analysis: a case study of biologics for rheumatoid arthritis, conceptual and technical challenges in network meta-analysis, questionable assumptions hampered interpretation of a network meta-analysis of primary care depression treatments., effects of study precision and risk of bias in networks of interventions: a network meta-epidemiological study., a critical review of methods for the assessment of patient-level interactions in individual participant data meta-analysis of randomized trials, and guidance for practitioners., combining randomized and non‐randomized evidence in network meta‐analysis, 27 references, borrowing strength from external trials in a meta-analysis., the results of direct and indirect treatment comparisons in meta-analysis of randomized controlled trials., combination of direct and indirect evidence in mixed treatment comparisons, indirect comparisons of competing interventions., health outcomes associated with various antihypertensive therapies used as first-line agents: a network meta-analysis., network meta‐analysis for indirect treatment comparisons, multiparameter evidence synthesis in epidemiology and medical decision‐making: current approaches, evaluation of networks of randomized trials, assessing evidence inconsistency in mixed treatment comparisons, fluoride gels for preventing dental caries in children and adolescents., related papers.
Showing 1 through 3 of 0 Related Papers

Published in Journal of Clinical Epidemiology 2009
G. Salanti V. Marinho J. Higgins
- Open access
- Published: 27 June 2011
Network meta-analysis-highly attractive but more methodological research is needed
- Tianjing Li 1 ,
- Milo A Puhan 1 ,
- Swaroop S Vedula 1 ,
- Sonal Singh 2 ,
- Kay Dickersin 1 &
The Ad Hoc Network Meta-analysis Methods Meeting Working Group
BMC Medicine volume 9 , Article number: 79 ( 2011 ) Cite this article
101k Accesses
271 Citations
97 Altmetric
Metrics details
Network meta-analysis, in the context of a systematic review, is a meta-analysis in which multiple treatments (that is, three or more) are being compared using both direct comparisons of interventions within randomized controlled trials and indirect comparisons across trials based on a common comparator. To ensure validity of findings from network meta-analyses, the systematic review must be designed rigorously and conducted carefully. Aspects of designing and conducting a systematic review for network meta-analysis include defining the review question, specifying eligibility criteria, searching for and selecting studies, assessing risk of bias and quality of evidence, conducting a network meta-analysis, interpreting and reporting findings. This commentary summarizes the methodologic challenges and research opportunities for network meta-analysis relevant to each aspect of the systematic review process based on discussions at a network meta-analysis methodology meeting we hosted in May 2010 at the Johns Hopkins Bloomberg School of Public Health. Since this commentary reflects the discussion at that meeting, it is not intended to provide an overview of the field.
Peer Review reports
Introduction
Systematic reviews use explicit, pre-specified methods to identify, appraise, and synthesize all available evidence related to a clinical question. When appropriate, systematic reviews may include a meta-analysis, that is, the statistical combination of results from two or more separate studies. Some systematic reviews compare only two interventions, in which a conventional pair-wise meta-analysis may be conducted, while others examine the comparative effectiveness of many or all available interventions for a given condition. When the comparative effectiveness of a range of interventions is of interest, appropriate statistical methodology must be used for analysis.
Also called mixed treatments comparison or multiple treatments comparison meta-analysis, network meta-analysis expands the scope of a conventional pair-wise meta-analysis by analyzing simultaneously both direct comparisons of interventions within randomized controlled trials (RCTs) and indirect comparisons across trials based on a common comparator (e.g., placebo or some standard treatment) [ 1 – 5 ]. In the simplest case, one may be interested in comparing two interventions A and C. Indirect evidence can be obtained from RCTs of either A or C versus a common comparator B (Figure 1 ), keeping intact the randomized comparisons within the RCTs [ 1 – 5 ]. When both direct and indirect evidence are available, the two sources of information can be combined as a weighted average when appropriate. Data structure of this type can be extended to k-comparisons to facilitate simultaneous inference regarding all available treatments, and to provide evidence for selecting the best of several treatment options. Many assumptions behind network meta-analysis methods appear to be similar to those made in standard pair-wise meta-analysis [ 6 ]. But as for a conventional pair-wise meta-analysis, the methodology for network meta-analysis must be carefully developed and rigorously evaluated before the technique is applied widely.
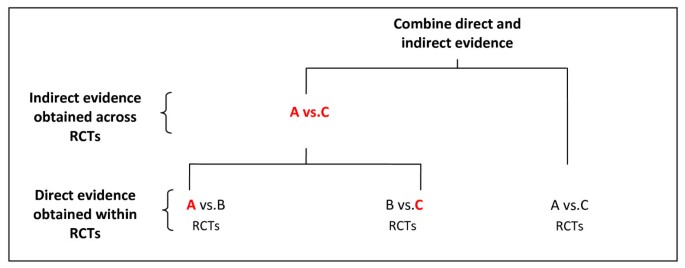
Illustration of a network meta-analysis that combines direct evidence obtained within RCTs (A vs. B, B vs. C and A vs. C), and indirect evidence obtained across RCTs through a common comparator (A vs. B and B vs. C) .
Despite a recent flurry of publications related to network meta-analyses [ 7 ], only a handful of articles have focused on key methodological issues and most of these have covered statistical approaches [ 2 – 4 , 8 – 16 ]. In May 2010, we hosted a meeting on network meta-analysis methodology at the Johns Hopkins Bloomberg School of Public Health. Vibrant discussions over the course of the meeting led us to identify major methodological questions concerning network meta-analysis and to propose a research agenda for the future. This article reflects discussion at the meeting and is not intended to provide an overview of the entire field.
Using statistical methods to combine findings from individual studies in a systematic review can provide useful information for clinical decision-making. To minimize error and ensure validity of findings from meta-analyses, the systematic review, whether it involves a standard, pair-wise meta-analysis or a network meta-analysis, must be designed rigorously and conducted carefully. Aspects of designing and conducting the systematic review include defining the review question, specifying eligibility criteria, searching for and selecting studies, assessing risk of bias and quality of evidence, conducting a meta-analysis, and interpreting and reporting findings [ 6 ]. The following sections discuss methodologic challenges and research opportunities for network meta-analysis relevant to each aspect of the systematic review process.
Define the review question and eligibility criteria
A well-formulated, clearly defined, answerable research question guides the eligibility criteria and the overall research protocol. Eligibility criteria combine aspects of the clinical question (e.g., Population, Interventions, Comparisons, and Outcomes) and specifications of the types of studies that have addressed this question [ 6 ]. Although the questions asked in pair-wise meta-analysis and network meta-analysis on a topic are different, the same interventions and comparisons may be examined, and these may be defined broadly or narrowly in both. For example, in both cases, one would want to define whether both drugs and behavioral interventions would be included, and if so, which ones. One would also want to define whether a different dose or regimen of the same treatment should be considered as the same or separate interventions.
Different specification of eligibility criteria may result in differences in the structure or extent of a network, leading to discrepant findings for network meta-analyses on the same topic. This is because different combinations of direct and indirect evidence, some independent and some overlapping, contribute to the comparisons and estimates of treatment effect [ 3 , 5 ]. Certain interventions, for example, interventions that are no longer in use, or placebos, may not be of primary interest but may be included in the network meta-analysis if they provide information concerning the interventions of interest through indirect comparisons. In a recent example, discordant conclusions were drawn from two systematic reviews that utilized direct and indirect evidence regarding the comparative effectiveness of second generation anti-depressants for major depression disorder [ 17 , 18 ]. One reason for the discrepancy was the difference in how the networks were defined [ 17 – 19 ]. One systematic review did not include placebo-controlled trials [ 17 ]. It is currently not possible to make general statements on the impact that different eligibility criteria may have on the validity of findings from a network meta-analysis.
Eligibility criteria in a review of harms may be different from a review of effectiveness because there might be limited data related to harm or adverse effects in a trial [ 20 ]. Methodologic research is needed to establish the role of non-randomized studies within a network meta-analysis evaluating harms associated with interventions.
Search for and select studies
To ensure that all relevant studies are identified, the network meta-analyst could search de novo for all relevant studies, but this would waste valuable resources if good systematic reviews with comprehensive searches already exist. To conserve valuable resources, one might consider using data identified through existing high quality systematic reviews of relevant pair-wise treatment comparisons provided the searches in the existing reviews are up-to-date. Empirical research is needed on the trade-offs associated with the two approaches to identify trials for a network meta-analysis. Such work will provide guidance for the network meta-analyst in choosing between conducting a new, comprehensive search or using existing searches.
As it is the case with a conventional pair-wise meta-analysis, the validity of findings from a network meta-analysis depends upon whether all eligible trials were identified and included in the analysis. Regardless of whether one conducts de novo searches or depends on existing systematic reviews, including a non-random or selective subset of all eligible trials in the analysis may introduce selection bias in the treatment effect estimates. Various forms of reporting biases have been identified in the literature [ 21 ]. As a consequence of reporting biases, for example, the network meta-analyst may fail to identify certain trials or when trials are identified, fail to retrieve data on outcomes relevant for analysis. One way that certain reporting biases are addressed is by conducting a search of multiple data sources for trial data. The various data sources that may be searched to retrieve trial data include published data, conference abstracts and other sources of grey literature, clinical trial registers, internal company reports, reviews of trials by regulatory agencies, and requesting trial investigators for individual patient data. Network meta-analysis involving both drug and non-drug interventions, for example, may be affected disproportionately if industry-sponsored trials are subject to greater reporting biases than other studies. Similarly, the internal validity of network meta-analysis of drug interventions may be affected if placebo-controlled trials are subject to greater reporting biases than active-controlled trials [ 22 ]. Methodological research is needed to examine the impact of various reporting biases and the use of multiple sources of trial data on the design, analysis, and findings from network meta-analyses.
Assess risk of bias and quality of evidence
The assessment of the risk of bias and its consideration in the network meta-analysis is far more challenging than in conventional meta-analysis. Risk of bias refers to the problems with the design and execution of individual trials that raise questions about the validity of their findings [ 6 ]. A fundamental difference between a conventional pair-wise meta-analysis and network meta-analysis is that a conventional pair-wise meta-analysis yields only one pooled effect estimate whereas a network meta-analysis yields more than one pooled effect estimate. Thus, while bias in the effect estimate from any single trial affects a single pooled effect estimate in a conventional meta-analysis, it may affect several pooled effect estimates obtained in a network meta-analysis. For example (Figure 1 ), the risk of bias for trials contributing to the direct comparison within a network may be low (e.g., all A vs. C trials described adequate masking), but the risk of bias for trials contributing to the indirect comparison may be high (e.g., some A vs. B or B vs. C trials reported no masking). In addition, the risk of bias may differ across different regions within the network of interventions being examined. Future methodological research should address ways to deal with such variation in risk of bias between direct and indirect comparisons and across the network. Specifically, such research may examine the impact of risk of bias in an individual trial on the network meta-analytic effect estimates, identify the biases specific to the network meta-analysis context that need to be considered, develop methods to assess, summarize and present the variation in risk of bias across the network, and use empirical research to postulate guidance for network meta-analysts on incorporating bias assessments in statistical analyses. Finally, methodological research may also examine whether network meta-analysis offers a potential method for identifying and adjusting for biases within included trials [ 10 , 15 , 23 ].
Conduct quantitative evidence synthesis
Several statistical methods are being used to implement network meta-analysis, for example, the adjusted indirect comparison method with aggregate data, meta-regression, hierarchical models, and Bayesian methods [ 1 ]. Some approaches provide better flexibility than others in adjusting for covariates and in ranking multiple interventions. Most network meta-analyses in the current literature use a single approach and the comparative performance of different approaches has not been studied in detail. Future methodological studies may evaluate the utility and robustness of various statistical methods, and identify circumstances in which specific methods or models are more efficient and appropriate than others.
Factors such as the total number of trials in a network, number of trials with more than two comparison arms, heterogeneity (i.e., clinical, methodological, and statistical variability within direct and indirect comparisons), inconsistency (i.e., discrepancy between direct and indirect comparisons), and bias may influence effect estimates obtained from network meta-analyses. Heterogeneity, inconsistency, and bias may propagate through a network of trials, and may affect the estimates differentially across regions of the network. A range of methods to detect, quantify and deal with heterogeneity, inconsistency, and bias has been proposed [ 10 – 12 , 15 , 23 ]. Evaluating the performance of the different methods, through simulations and empirical studies, is critical before they become widely available.
Most network meta-analyses to date use WinBUGs software, which is limited in functionality and accessibility to the non-statistician. New software is needed that balances user-friendliness with statistical sophistication and provide built-in methodological guidance. In addition, new software should be able to handle in a coherent manner different types of outcomes (e.g., continuous outcomes, binary outcomes), multiple outcomes, outcomes at different follow-up times and simultaneously carry out pair-wise and network meta-analysis.
With availability of the new, easy to use software, concerns arise about network meta-analysis being undertaken and implemented inappropriately. Thus, systematic reviewers should be educated to identify potential research questions where network meta-analysis may be appropriate, and where it is not, including the situation where the evidence is sparse.
Interpret results and report findings
Presenting and communicating complex findings from a network meta-analysis in an accessible and understandable format is challenging. It is critical to report all pair-wise effect estimates together with the associated confidence or credible intervals, depending on the statistical model used (i.e., frequentist or Bayesian model). Probability statements could be made about the effectiveness of each treatment [ 24 ]. For example, for each treatment, one can calculate the probability that the treatment is the best, second best, or third best among all treatments. Such probability statements should be interpreted carefully since the difference between treatments might be small and not clinically meaningful.
In addition to the estimates of treatment effects, uncertainty, clinical and methodological characteristics, and potential biases within included trials must be conveyed. A careful assessment of the body of evidence and a thoughtful discussion of the potential impact of trial-specific biases on the effect estimates in a network meta-analysis can maximize transparency and avoid errors in interpretation. Using the hypothetical example described in a preceding section, if the preponderance of evidence within the network is constituted by trials that did not report masking, interpreting effect estimates from a network meta-analysis of such trials should be tempered by a discussion on the impact of potential bias due to inadequate masking. Values and preferences from potential evidence users should be considered in interpretation. Guidelines may be developed, based on methodological research, to establish standards for reporting network meta-analyses. Although a recent survey identified nearly 100 published network meta-analyses published between 2000 and 2007 [ 7 ], many peer reviewers are relatively uneducated in these methods. Guidance may be developed to aid rigorous peer review of findings from network meta-analyses submitted to medical journals.
Conclusions
This commentary summarizes the methodologic challenges and areas of research for network meta-analysis relevant to each aspect of the systematic review process based on discussions at a meeting. It is not intended to provide a comprehensive overview of the field. Network meta-analysis holds promise to provide evidence on comparative effectiveness that is valuable for clinical decision-making because it allows comparisons of interventions that may not have been directly compared in head-to-head trials. Collaborative efforts between epidemiologists, statisticians, clinicians and others are necessary for developing, implementing and evaluating methods for network meta-analysis. The extent to which the medical community accepts network meta-analysis will depend on how convincingly methodological research demonstrates the validity of the evidence and its ease of interpretation for decision-makers.
Glenny AM, Altman DG, Song F, Sakarovitch C, Deeks JJ, D'Amico R, et al: Indirect comparisons of competing interventions. Health Technol Assess. 2005, 9 (26): 1-134.
Article CAS PubMed Google Scholar
Caldwell DM, Ades AE, Higgins JP: Simultaneous comparison of multiple treatments: combining direct and indirect evidence. BMJ. 2005, 331 (7521): 897-900. 10.1136/bmj.331.7521.897.
Article PubMed PubMed Central Google Scholar
Lu G, Ades AE: Combination of direct and indirect evidence in mixed treatment comparisons. Stat Med. 2004, 23 (20): 3105-24. 10.1002/sim.1875.
Higgins JP, Whitehead A: Borrowing strength from external trials in a meta-analysis. Stat Med. 1996, 15 (24): 2733-49. 10.1002/(SICI)1097-0258(19961230)15:24<2733::AID-SIM562>3.0.CO;2-0.
Lumley T: Network meta-analysis for indirect treatment comparisons. Stat Med. 2002, 21 (16): 2313-24. 10.1002/sim.1201.
Article PubMed Google Scholar
Higgins JPT, Green S, (eds): Cochrane Handbook for Systematic Reviews of Interventions Version 5.1.0. 2011, The Cochrane Collaboration, [updated March 2011], [ http://www.cochrane-handbook.org ]
Google Scholar
Song F, Loke YK, Walsh T, Glenny AM, Eastwood AJ, Altman DG: Methodological problems in the use of indirect comparisons for evaluating healthcare interventions: survey of published systematic reviews. BMJ. 2009, 338: b1147-10.1136/bmj.b1147.
Ades AE: A chain of evidence with mixed comparisons: models for multi-parameter synthesis and consistency of evidence. Stat Med. 2003, 22 (19): 2995-3016. 10.1002/sim.1566.
Caldwell DM, Welton NJ, Ades AE: Mixed treatment comparison analysis provides internally coherent treatment effect estimates based on overviews of reviews and can reveal inconsistency. J Clin Epidemiol. 2010, 63 (8): 875-82. 10.1016/j.jclinepi.2009.08.025.
Dias S, Welton NJ, Ades AE: Study designs to detect sponsorship and other biases in systematic reviews. J Clin Epidemiol. 2010, 63 (6): 587-8. 10.1016/j.jclinepi.2010.01.005.
Dias S, Welton NJ, Caldwell DM, Ades AE: Checking consistency in mixed treatment comparison meta-analysis. Stat Med. 2010, 29 (7-8): 932-44. 10.1002/sim.3767.
Lu G, Ades AE: Assessing evidence inconsistency in mixed treatment comparisons. J Am Stat Assoc. 2006, 101 (474): 447-59. 10.1198/016214505000001302.
Article CAS Google Scholar
Lu G, Ades AE, Sutton AJ, Cooper NJ, Briggs AH, Caldwell DM: Meta-analysis of mixed treatment comparisons at multiple follow-up times. Stat Med. 2007, 26 (20): 3681-99. 10.1002/sim.2831.
Hasselblad V: Meta-analysis of multitreatment studies. Med Decis Making. 1998, 18 (1): 37-43. 10.1177/0272989X9801800110.
Salanti G, Higgins JP, Ades AE, Ioannidis JP: Evaluation of networks of randomized trials. Stat Methods Med Res. 2008, 17 (3): 279-301.
Salanti G, Marinho V, Higgins JP: A case study of multiple-treatments meta-analysis demonstrates that covariates should be considered. J Clin Epidemiol. 2009, 62 (8): 857-64. 10.1016/j.jclinepi.2008.10.001.
Cipriani A, Furukawa TA, Salanti G, Geddes JR, Higgins JP, Churchill R, et al: Comparative efficacy and acceptability of 12 new-generation antidepressants: a multiple-treatments meta-analysis. Lancet. 2009, 373 (9665): 746-58. 10.1016/S0140-6736(09)60046-5.
Gartlehner G, Gaynes BN, Hansen RA, Thieda P, DeVeaugh-Geiss A, Krebs EE, et al: Comparative benefits and harms of second-generation antidepressants: background paper for the American College of Physicians. Ann Intern Med. 2008, 149 (10): 734-50.
Ioannidis JP: Ranking antidepressants. Lancet. 2009, 373 (9677): 1759-60.
Chou R, Aronson N, Atkins D, Ismaila AS, Santaguida P, Smith DH, et al: AHRQ series paper 4: assessing harms when comparing medical interventions: AHRQ and the effective health-care program. J Clin Epidemiol. 2010, 63 (5): 502-12. 10.1016/j.jclinepi.2008.06.007.
Song F, Parekh S, Hooper L, Loke YK, Ryder J, Sutton AJ, et al: Dissemination and publication of research findings: an updated review of related biases. Health Technol Assess. 2010, 14 (8): 1-193.
Article Google Scholar
Rising K, Bacchetti P, Bero L: Reporting bias in drug trials submitted to the Food and Drug Administration: review of publication and presentation. PLoS Med. 2008, 5 (11): e217-10.1371/journal.pmed.0050217. discussion e217. Erratum in: PLoS Med. 2009;6(1)
Song F, Harvey I, Lilford R: Adjusted indirect comparison may be less biased than direct comparison for evaluating new pharmaceutical interventions. J Clin Epidemiol. 2008, 61 (5): 455-63. 10.1016/j.jclinepi.2007.06.006.
Salanti G, Ades AE, Ioannidis JP: Graphical methods and numerical summaries for presenting results from multiple-treatment meta-analysis: an overview and tutorial. J Clin Epidemiol. 2011, 64 (2): 163-71. 10.1016/j.jclinepi.2010.03.016.
Pre-publication history
The pre-publication history for this paper can be accessed here: http://www.biomedcentral.com/1741-7015/9/79/prepub
Download references
Acknowledgements and Funding
The meeting was sponsored by Grant 1 RC1 EY020140-01, National Eye Institute, National Institutes of Health, United States. The sponsor had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; and preparation, review, or approval of the manuscript.
Ad Hoc Network Meta-analysis Methods Meeting Working Group (alphabetically ordered by last name): Chris Cameron, Canadian Agency for Drugs and Technologies in Health; Kay Dickersin, Johns Hopkins Bloomberg School of Public Health; Steven N. Goodman, Johns Hopkins Medical Institutions; Tianjing Li, Johns Hopkins Bloomberg School of Public Health; Edward Mills, University of Ottawa; David Musch, University of Michigan; Milo A. Puhan, Johns Hopkins Bloomberg School of Public Health; Gerben ter Riet, University of Amsterdam; Karen Robinson, Johns Hopkins Medical Institutions; Christopher Schmid, Tufts University; Sonal Singh, Johns Hopkins Medical Institutions; Fujian Song, University of East Anglia; Kristian Thorlund, McMaster University; Thomas Trikalinos, Tufts University; Swaroop S. Vedula, Johns Hopkins Bloomberg School of Public Health.
Author information
Authors and affiliations.
Johns Hopkins Bloomberg School of Public Health, 615 N. Wolfe Street, Mail Room W5010, Baltimore, Maryland, 21212, USA
Tianjing Li, Milo A Puhan, Swaroop S Vedula & Kay Dickersin
Johns Hopkins Medical Institutions, 1830 E. Monument St, Suite 8063, Baltimore, Maryland, 21212, USA
Sonal Singh
You can also search for this author in PubMed Google Scholar
Corresponding author
Correspondence to Kay Dickersin .
Additional information
Competing interests.
TL, MP, and KD reported receiving salary support though a grant to the Johns Hopkins Bloomberg School of Public Health from the National Eye Institute, National Institutes of Health to conduct the study and to write the manuscript. The grant also supports TL, MP, and KD to travel to meetings related to the study. SV and SS declared that they have no financial or non-financial competing interests.
Authors' contributions
TL has full access to all of the transcriptions of meeting discussions, and takes responsibility for the integrity of the manuscript. TL, MP, SV, SS, and KD participated in the study conception, design, and analysis, and interpretation of findings. TL, MP, and SV drafted the manuscript. SS and KD reviewed and edited the manuscript for important intellectual content. KD obtained funding for this study, and supervised the work. TL and KD also provided administrative and material support. Meeting contributors either presented their current research and ideas, or provided insightful comments during the meeting, and sent constructive feedback to the manuscript draft. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Authors’ original file for figure 1
Rights and permissions.
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Reprints and permissions
About this article
Cite this article.
Li, T., Puhan, M.A., Vedula, S.S. et al. Network meta-analysis-highly attractive but more methodological research is needed. BMC Med 9 , 79 (2011). https://doi.org/10.1186/1741-7015-9-79
Download citation
Received : 04 January 2011
Accepted : 27 June 2011
Published : 27 June 2011
DOI : https://doi.org/10.1186/1741-7015-9-79
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Indirect Comparison
- Major Depression Disorder
- Mixed Treatment Comparison
- Common Comparator
- Pool Effect Estimate
BMC Medicine
ISSN: 1741-7015
- General enquiries: [email protected]
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- Open access
- Published: 29 August 2024
A targetable type III immune response with increase of IL-17A expressing CD4 + T cells is associated with immunotherapy-induced toxicity in melanoma
- Florentia Dimitriou ORCID: orcid.org/0000-0003-4529-3372 1 , 2 ,
- Phil F. Cheng ORCID: orcid.org/0000-0003-2940-006X 1 , 2 , 3 ,
- Annalisa Saltari 1 , 2 ,
- Katrin Schaper-Gerhardt 4 , 5 ,
- Ramon Staeger ORCID: orcid.org/0000-0002-9283-9596 1 , 2 ,
- Veronika Haunerdinger 1 , 2 ,
- Federica Sella ORCID: orcid.org/0000-0002-0805-0905 1 , 2 ,
- Aizhan Tastanova ORCID: orcid.org/0000-0003-1120-5610 1 , 2 ,
- Christian Urban 6 ,
- Susanne Dettwiler 7 ,
- Daniela Mihic-Probst 2 , 7 ,
- Christian M. Matter 8 ,
- Olivier Michielin 3 ,
- Ralf Gutzmer ORCID: orcid.org/0000-0001-7921-2820 4 ,
- Georgina V. Long ORCID: orcid.org/0000-0001-8894-3545 9 , 10 , 11 ,
- Burkhard Becher ORCID: orcid.org/0000-0002-1541-7867 12 ,
- Mitchell P. Levesque ORCID: orcid.org/0000-0001-5902-9420 1 , 2 na1 &
- Reinhard Dummer ORCID: orcid.org/0000-0002-2279-6906 1 , 2 na1
Nature Cancer ( 2024 ) Cite this article
1 Altmetric
Metrics details
- Cancer immunotherapy
Immune checkpoint inhibitors are standard-of-care for the treatment of advanced melanoma, but their use is limited by immune-related adverse events. Proteomic analyses and multiplex cytokine and chemokine assays from serum at baseline and at the adverse event onset indicated aberrant T cell activity with differential expression of type I and III immune signatures. This was in line with the finding of an increase in the proportion of CD4 + T cells with IL-17A expression at the adverse event onset in the peripheral blood using flow cytometry. Multiplex immunohistochemistry and spatial transcriptomics on immunotherapy-induced skin rash and colitis showed an increase in the proportion of CD4 + T cells with IL-17A expression. Anti-IL-17A was administered in two patients with mild myocarditis, colitis and skin rash with resolution of the adverse events. This study highlights the potential role of type III CD4 + T cells in adverse event development and provides proof-of-principle evidence for a clinical trial using anti-IL-17A for treating adverse events.
Similar content being viewed by others
Anti-TIGIT antibody improves PD-L1 blockade through myeloid and T reg cells
Distinct molecular and immune hallmarks of inflammatory arthritis induced by immune checkpoint inhibitors for cancer therapy
A T cell inflammatory phenotype is associated with autoimmune toxicity of the PI3K inhibitor duvelisib in chronic lymphocytic leukemia
Immune checkpoint inhibitors (ICIs) block the immunosuppressive milieu that promotes tumorigenesis and tumor progression and are therefore established for the treatment of multiple malignancies, including melanoma 1 . This disturbance of the homeostatic mechanisms of immune tolerance may lead to various toxicities, referred to as immune-related adverse events (irAEs) 2 . Severe (grade ≥3) irAEs leading to systemic corticosteroid administration and hospitalization have been reported in 17% of patients treated with single-agent anti-PD1 (ref. 3 ); they occur more frequently when ICIs are combined 4 . Although these irAEs are generally manageable with the administration of systemic corticosteroids or other immunomodulatory agents 5 , 6 , they represent a significant source of morbidity and can be rarely associated with long-term functional organ impairment 7 or even fatality 8 . Hence, treatment strategies to mitigate irAEs without compromising antitumor immunity are urgently needed.
A comprehensive mechanistic understanding of the development of irAEs is currently missing. Some of the main components involved in irAE occurrence include autoreactive and cross-reactive T cells and B cells against tumor-specific antigens 9 , 10 , 11 , complement-mediated inflammation 12 , 13 and expansion of pre-existing autoantibodies produced by B cells 14 , 15 . Ultimately, irAEs are linked to the mechanism of action of ICIs, which results in alterations of the systemic immunity; CTLA4 inhibition leads to increased priming and activation of antigen-specific T cells, whereas inhibition of PD1 signaling stimulates oligo-clonal expansion of T cells at the tumor site 14 . Activated T cells are primed towards a certain lineage, which is regulated by stimulating and inhibiting cytokines. To comprehensively describe this helper T cell (T H ) plasticity, three types of responses that are based on the primary engaged target cells have been proposed: type I is triggered by intracellular pathogens with the production of IFNγ and GM-CSF; type II supports the development of cellular immunity with the production of IL-4, IL-5 and IL-13; and type III is initiated at barrier sites, such as the skin, gut and other mucosal tissue 16 . Critical cytokines of the type III T H cells include IL-17 and IL-22, which are expressed throughout the stromal and immune compartment, and their dysregulation has been implicated in various diseases of barrier tissues 17 , 18 , 19 . Type III cytokines, such as IL-6, have also been linked to the occurrence of irAEs in patients treated for melanoma 20 , 21 . These data, alongside the successful use of cytokine-blocking treatments in the management of corticosteroid-refractory irAEs 22 , 23 , raise the possibility of additional therapeutic strategies in addition to the standard use of corticosteroids. Based on these findings, we designed a translational study to comprehensively analyze the immune biology of irAEs and propose possible treatment strategies in addition to the standard administration of systemic corticosteroids.
Increase of cytokines and chemokines in patients with severe irAEs
We systematically analyzed serum proteome and circulating chemokines and cytokines from peripheral blood collected at baseline (0–29 days before therapy initiation) and at the irAEs onset or at the second to fifth infusion (in patients without any irAEs), in patients with advanced melanoma treated with anti-PD1-based ICIs, including single-agent anti-PD1 (pembrolizumab and nivolumab) or in combination with anti-CTLA4 (ipilimumab) in a discovery, verification and validation cohort (Fig. 1 ). As an initial approach to investigate the correlation of specific cytokines and chemokines with the onset of irAEs, we used a discovery cohort of nine patients with stage IV melanoma and severe (grade ≥3) irAEs during the treatment course of ipilimumab and nivolumab. This discovery cohort was used as an exploratory cohort to investigate the correlation of specific cytokines and chemokines with the onset of the irAEs. For this purpose, serum samples that were collected at the baseline served as controls to investigate the change of specific cytokines and chemokines in samples collected at the onset of the irAEs. Based on the results of this analysis, a study protocol was established for prospective sample collection in a main study cohort, in which patients with irAEs (cases) were compared to control patients without irAEs.

Overview of the patients included in the study and downstream analyses that were performed. Created with BioRender.com .
Serum samples were analyzed using a panel of 33 cytokines and chemokines with the U-PLEX Assay Platform (Meso Scale Diagnostics (MSD)). Patients were retrospectively selected according to the availability of serum samples at baseline and at the onset of an irAE from an initial cohort of 53 patients with similar treatment and irAE characteristics. Baseline characteristics are summarized in Supplementary Table 1 . Seven patients (78%) had multiple, concurrent irAEs that included ≥2 systemic organs, and the most common irAEs included immune-related colitis (5 out of 9, 56%), ir-hepatitis (3 out of 9, 33%) and skin rash (3 out of 9, 33%). Initial analyses of the serum cytokines and chemokines at baseline using a multiplex chemokine/cytokine assay revealed that most cytokines and chemokines were expressed at low levels, which increased at irAE onset (Extended Data Fig. 1a,b ). In a Wilcoxon rank sum test analysis for the significant cytokines and chemokines, IL-17A showed the highest log 2 (fold change) from baseline to irAE onset (3.81, P < 0.01, respectively); of note, the IL-17A elevation at irAE onset was overall consistent. Further cytokines and chemokines included IFNγ (3.77, P < 0.01), CXCL9 (3.02, P < 0.01), CXCL11 (2.3, P < 0.01), IL-10 (2.28, P < 0.01), IL-21 (2.06, P < 0.01), CXCL10 (1.91, P < 0.01) and TNF (1.79, P < 0.01).
Proteomic analyses of serum proteins at the onset of irAEs
A proteomic analysis based on the proximity extension assay technology and the use of Olink Explore 384 Inflammation panel on serum samples prospectively collected at baseline and the onset of irAEs or at the second to fifth infusion (in patients without irAEs) was implemented in a main study cohort of 73 patients. Patient characteristics are described in Supplementary Table 1 . Overall, 49 out of 73 patients (67%) developed irAEs, 29 out of 73 (40%) had multiple (≥2) concurrent irAEs and 34 out of 73 (47%) had severe (grade ≥3) irAEs. The median time to irAEs onset was 27 days (range, 1–91 days). The overall response rate was 49%.
Patients with irAEs had higher log 2 (fold change) in several proteins of this panel than those without any irAEs (Fig. 2a ). For the differential protein analysis between the two time points, namely TP 0 (baseline) and TP 1 (irAEs onset or second to fifth infusion), we considered a fold change of two as a relevant change, which corresponds to a log 2 (fold change) of >1, with an adjusted P value of ≤0.05. Using the Wilcoxon rank sum test, four proteins showed a significant differential expression from TP 0 to TP 1 in patients with irAEs; these included CXCL9, CXCL10, IFNγ and IL-10 (Fig. 2b ). None of these proteins showed any significant change in patients without irAEs. The same test was performed for the differential protein analysis in patients with multiple (≥2) irAEs compared to those with single irAEs (Extended Data Fig. 2a ) as well as those with severe (grade ≥3) irAEs compared to those with non-severe (grade 1–2) irAEs (Extended Data Fig. 2b ). Notably, the differential expression of the significant proteins in patients with multiple (≥2) irAEs and those with severe (grade ≥3) irAEs was similar and included eight overlapping proteins: CXCL9, CXCL10, IL-10, IFNγ, GZMA, FABP1, TNF and SULT2A1. To further characterize the significant proteins that were differentially expressed between TP 0 and TP 1 , and to analyze these results in one statistical model, we identified proteins that significantly changed over time and between the two patient groups, namely those with and without irAEs, using a linear mixed-effect model for the time and the occurrence of the irAEs. Other covariates, including age, sex and treatment type, were integrated into the model, and each patient was added as a random effect. Using this statistical model, 11 proteins showed the highest differential expression from baseline to the irAE onset in patients with irAEs (all P < 0.01; Fig. 2c ). These included (1) CXCL9, which has chemotactic functions and regulates the immune cell migration, differentiation and activation 24 ; (2) IL-17A, a pro-inflammatory cytokine produced by activated T cells 25 ; (3) IL-15, a pleiotropic inflammatory cytokine that regulates the homeostasis of both innate and adaptive immune cells 26 ; (4) keratin 19 (KRT19), a protein-coding gene that encodes the protein CYFRA21.1, a member of the keratin family 27 ; (5) CD276, a type I transmembrane protein that suppresses T cell activation and proliferation in non-malignant tissues 28 ; (6) IL2RB, which is involved in T cell-mediated immune responses and increases proliferation of CD8 + effector T cells 29 ; (7) CD70, which has an important role in the regulation of the immune system activation 30 ; (8) follistatin-related protein 3 (FSTL3), a secreted glycoprotein of the follistatin-module-protein family 31 ; (9) Fms-related tyrosine kinase 3 ligand (FLT3LG), which stimulates the proliferation and differentiation of various blood cell progenitors, including the growth of dendritic cells 32 ; (10) beta-1,4-galactosyltransferase 1 (B4GALT1), which is overexpressed in pathological processes, including inflammation and proliferation of cancer cells 33 ; and (11) placental growth factor, a member of the vascular endothelial growth factor (VEGF) sub-family, a key molecule in angiogenesis and vasculogenesis 34 . Of note, some of these proteins also increased over time in patients without irAEs, but these changes were not significant in the linear mixed-effect model (Fig. 2c ). None of these proteins was predictive for the occurrence of irAEs at baseline.

a , Heatmap of log 2 (FC) of the 384 serum proteins in n = 73 patients with and without irAEs in the main study cohort b , Volcano plot with the differential expressed proteins at baseline (TP 0 ) and at irAE onset (in patients with irAEs) or at the second to fifth infusion (in patients without irAEs) (TP 1 ) in the main study cohort ( n = 73 patients). Significant difference was determined by a two-sided Wilcoxon rank sum test, and P values were adjusted by the Benjamini–Hochberg method. FDR, false discovery rate; FC, fold change. c , Linear mixed-effects regression analysis of the 11 significant proteins for the two time points (TP 0 and TP 1 ) in n = 73 patients with and without irAEs. Data are represented by the estimated marginal means with 95% confidence intervals. The y axis represents the Olink assay value (NPX). P values are adjusted using the Tukey method.
Source data
Next, the differential protein expression was analyzed according to the treatment response; using the Wilcoxon rank sum test, responders and non-responders showed a significant increase in specific proteins from TP 0 to TP 1 , which included CXCL9, IL-10 and IFNγ in responders, and CXCL9 and CXCL10 in non-responders (false discovery rate-adjusted P ≤ 0.05 and log 2 (fold change) > 1; Extended Data Fig. 2c ). To further analyze these results in one statistical model, a linear mixed-effect model for the time, treatment response and occurrence of the irAEs was used. This statistical model was adjusted for age and sex. None of the 11 proteins that showed the highest expression in patients with irAEs was significant for the type of treatment response (Extended Data Fig. 2d ). Overall, this analysis shows that specific proteins are significantly upregulated at the time of the irAE onset in patients with irAEs.
Increase of type I and III cytokines and chemokines at the onset of irAEs
A multiplex chemokine/cytokine assay with the selected panel of 33 cytokines and chemokines described above (MSD) was applied to further validate these results in the main study cohort. Of note, the MSD technology provides a multiplex assay of high sensitivity and broad dynamic range for investigating cytokines and chemokines using electrochemiluminescence, which allows for technical validation of the results with high accuracy 35 . Similar to the proteomic analysis, patients with irAEs showed higher log 2 (fold change) in several cytokines and chemokines than those without any irAEs (Fig. 3a ), as did patients with multiple (≥2) irAEs (Extended Data Fig. 3 ). Using the Wilcoxon rank sum test, seven cytokines and chemokines showed the highest log 2 (fold change) from TP 0 to TP 1 in patients with irAEs, including CXCL9, CXCL10, IL-17A, IL-12A, IL-10, IL-21 and IFNγ (false discovery rate-adjusted P ≤ 0.05 and log 2 (fold change) > 1; Fig. 3b ).

a , Heatmap of log 2 (fold change) of the serum cytokines and chemokines per patient in the multiplex MSD assay at the baseline (TP 0 ) and at the irAE onset (in patients with irAEs) or at the second to fifth infusion (in patients without irAEs) (TP 1 ) in the main study cohort ( n = 73 patients). b , Volcano plot with the differential expressed proteins at TP 0 and TP 1 in patients with and without irAEs in the multiplex MSD assay in the main study cohort ( n = 73 patients). Significant difference was determined by a two-sided Wilcoxon rank sum test, and P values were adjusted by the Benjamini–Hochberg method. c , Heatmap of log 2 (fold change) of the serum cytokines and chemokines per patient in the multiplex MSD assay at TP 0 and TP 1 in the external validation cohort ( n = 81 patients). d , Receiver operating characteristic curve analysis of the five cytokine and chemokine panels that were significantly differentially expressed in the main study cohort ( n = 73 patients) for the association with the irAE onset in the external validation cohort ( n = 81 patients). AUC, area under the curve.
The analytical validation of these results was performed in an independent external validation cohort of n = 81 patients using a similar multiplex cytokine–chemokine assay (MSD). The baseline characteristics of this cohort are summarized in Supplementary Table 1 . In line with the previous observations, most cytokines and chemokines increased from TP 0 to TP 1 (Fig. 3c ). Of note, CXCL9 and IL-21 were not included in the multiplex cytokine–chemokine assay of the external validation cohort. To analyze the performance of the five (out of seven) cytokines and chemokines that showed the highest log 2 (fold change) in the verification phase of the study, namely CXCL10, IL-17A, IL-10, IL-12A and IFNγ, we performed a receiver operating characteristics curve analysis and found an area under the curve of 0.69 (95% CI, 0.58–0.81) (Fig. 3d ) 36 . Overall, these results allow for a reliable association of signaling activities at the onset of irAEs and further identification of possible therapeutic biomarkers.
The cellular source of significant cytokines and chemokines
Single-cell RNA sequencing (scRNA-seq) analysis of peripheral blood mononuclear cells (PBMCs) collected at TP 0 and TP 1 was performed to identify the cellular source of the cytokines and chemokines involved in these conditions. Taking into consideration the non-specific changes in the cytokine and chemokine profiles in responders and non-responders, PBMCs from patients with similar responses to ICIs were selected to eliminate any potential bias in the interpretation of the results. As such, PBMCs from six patients treated with ipilimumab and nivolumab with a partial response to the systemic treatment as the best overall response were analyzed using scRNA-seq. Three patients had grade ≥3 irAEs and three patients did not develop any irAEs during the treatment course. The single-cell data were analyzed using the Seurat R package, and cells were typed with SingleR using the Monaco Immune Data reference ( GSE107011 ). Dimension reduction by principal component analysis (PCA), clustering by the Leiden algorithm and projection into 2D space by uniform manifold approximation and projection (UMAP) uncovered 24 distinct subpopulations from 117,702 cells. These cells were then evaluated, and eight major mononuclear lineages were encompassed, including B cells, CD4 + and CD8 + T cells, unspecified T cells, natural killer cells, monocytes, dendritic cells and progenitor cells (Fig. 4a ). Dot plot analysis for the significantly differentially expressed cytokines and chemokines of the multiplex MSD assay in the main immune cell types showed an increased average expression of the type I cytokines and chemokines from TP 0 to TP 1 in patients with irAEs (Fig. 4b ). The cellular source of the eight significant cytokines and chemokines that showed the highest log 2 (fold change) from the baseline to the irAE onset was then visualized using UMAP and cell typing (Fig. 4c–n ). In brief, CXCL9 and CXCL10 signals showed significant changes from TP 0 to TP 1 in patients with irAEs in monocytes and natural killer cells, whereas IFNγ was significantly expressed by CD4 + , CD8 + and natural killer cells from TP 0 to TP 1 in both patient groups. For IL-17A, no expression was detected.

a , UMAP plot, with overview of the main immune cell types for the above-mentioned conditions ( n = 6 patients). A total of 118,734 PBMCs were sequenced. NK, natural killer b , Dot plot with the average expression of the significantly differentially expressed cytokines and chemokines from the multiplex MSD assay in the main immune cell types in the scRNA-seq analysis at the two time points (TP 0 and TP 1 ) for the six patients. The size of the dot represents the percentage of cells that show expression and the color of the dot represents the average expression. c , d , UMAP for CXCL9 for TP 0 and TP 1 in patients with and without irAEs ( n = 6) ( c ) and violin plot for the CXCL9 signal in main cell types for TP 0 and TP 1 in patients with and without irAEs ( n = 6) ( d ). e , f , UMAP for CXCL10 for TP 0 and TP 1 in patients with and without irAEs ( n = 6) ( e ) and violin plot for the CXCL10 signal in main cell types for TP 0 and TP 1 in patients with and without irAEs ( n = 6) ( f ). g , h , UMAP for IFNγ for TP 0 and TP 1 in patients with and without irAEs ( n = 6) ( g ) and violin plot for the IFNγ signal in main cell types for TP 0 and TP 1 in patients with and without irAEs ( n = 6) ( h ). i , j , UMAP for IL-10 for TP 0 and TP 1 in patients with and without irAEs ( n = 6) ( i ) and violin plot for the IL-10 signal in main cell types for TP 0 and TP 1 in patients with and without irAEs ( n = 6) ( j ). k , l , UMAP for IL-12A for TP 0 and TP 1 in patients with and without irAEs ( n = 6) ( k ) and violin plot for the IL-12A signal in main cell types for TP 0 and TP 1 in patients with and without irAEs ( n = 6) ( l ). m , n , UMAP for IL-21 for TP 0 and TP 1 in patients with and without irAEs ( n = 6) ( m ) and violin plot for the IL-21 signal in main cell types for TP 0 and TP 1 in patients with and without irAEs ( n = 6) ( n ).
Each subpopulation was then analyzed at TP 0 and TP 1 . The UMAP for each of these conditions showed that the transcriptomic profile of the immune cells altered from TP 0 to TP 1 in patients with irAEs (Extended Data Fig. 4 ). Next, we specifically looked for the T cell subtypes, which were visualized using UMAP (Fig. 5a ). Differential gene expression analysis for the T cell subsets revealed a significant increase in STAT1 in all T cell subtypes in patients with irAEs and most T cell subtypes in patients without irAEs (Fig. 5b,c ). By contrast, STAT3 was significantly increased in T H 17 cells in patients with irAEs ( P < 0.05) (Fig. 5b,d ). Of note, STAT3 was significantly increased in terminal effector CD8 + T cells in patients without irAEs, which, alongside the existing literature involving the IL-10–IL-21–STAT3 pathway in the development of memory CD8 + T cells 37 , might imply a higher activation of this pathway in patients without irAEs that respond to the ICI treatment. This finding should be investigated more rigorously in future studies.

a , UMAP plot with overview of the T cell subtypes for the n = 6 patients. b , Heatmap with the significantly differentially expressed genes for each T cell subtype at TP 0 and TP 1 for patients with and without irAEs ( n = 6). c , Violin plot of the STAT1 gene for the T cell subtypes between TP 0 and TP 1 ( n = 6). STAT1 normalized counts between TP 0 and TP 1 were compared using a two-sided Wilcoxon rank sum test and P values adjusted by the Benjamini–Hochberg method. d , Violin plot of the STAT3 gene for the T cell subtypes between TP 0 and TP 1 ( n = 6). STAT3 normalized counts between TP 0 and TP 1 were compared using a two-sided Wilcoxon rank sum test and P values adjusted by the Benjamini–Hochberg method.
Increase of CD4 + T cells with IL-17A expression at the irAEs onset
Cytokine signaling activity at a single-cell mRNA expression level in peripheral blood has several limitations. To limit the impact on the interpretation of previous results from scRNA-seq data, we additionally performed immunoprofiling of the T cells that are able to produce IL-17A 38 , 39 using flow cytometry in the PBMCs collected at TP 0 and TP 1 from the six patients of the above-mentioned cohort. The purpose of this analysis was to reliably delineate the cellular source of the IL-17A production at the irAE onset. We reasoned that dysregulation of the IL-17A-producing cells contributes to the onset of irAEs and that their immune signature can be used as therapeutic targets in these patients.
The collected PBMCs from patients with and without irAEs from TP 0 and TP 1 were stimulated for intracellular cytokine staining. For the cell type identification, antibodies targeting CD4 + and CD8 + T cells, B cells and γδ T cells were included (Extended Data Fig. 5 and Extended Data Fig. 6a ). In the flow cytometry analysis, IL-17A production was only detectable in CD4 + T cells (Extended Data Fig. 6b ). The proportional analysis of the IL-17A-producing CD4 + T cells for the two time points revealed a significant increase between patients with and without irAEs at TP 1 ( P = 0.05), which was not present at TP 0 ( P = 0.2) (Extended Data Fig. 6c ). These results demonstrate that CD4 + T cells are able to produce IL-17A at the irAE onset in patients with irAEs, thus underlining their substantial role in the occurrence of irAEs.
T H 17-mediated inflammation in colitis and skin rash samples
We next reasoned that the increase of the IL-17A-producing cells in the peripheral blood reflected changes at the site of occurrence of the irAEs. To address this hypothesis, multiplex immunofluorescence analysis of immune-related colitis and lichenoid skin rash samples of the six patients with available PBMCs that were included in the scRNA-seq analysis was performed, aiming to further elucidate the immune cells involved in the irAEs and to validate the above-mentioned results at the site of the inflamed tissue. Of note, lichenoid skin rashes are the most common dermatologic irAEs in patients treated with ICIs 40 , whereas the incidence of immune-related colitis ranges between 8% and 27% 6 , with diarrhea being a common treatment-related irAE during treatment with ipilimumab and nivolumab treatment 41 . Formalin-fixed, paraffin-embedded (FFPE) skin biopsies from six patients with lichenoid skin rash during treatment with ipilimumab and nivolumab were stained for CD4, CD8, FoxP3, IL-17A, CD68 and pan-cytokeratin (panCK) and compared to normal skin from healthy, consenting individuals obtained through excess skin removal from plastic surgery (Fig. 6a–c ). The histopathology reports were reviewed by two independent pathologists. Annotation and quantification of the immune cells was performed with Akoya inForm software (v.2.6.0). Cell classification was based on the protein expression; CD4 + and IL-17A + cells were labeled as T H 17. A range of 2.3–24.9% cell proportion of IL-17A-producing cells was observed in all six skin rash biopsies compared to 0.3% T H 17 cells in the normal skin (Fig. 6d ). Regulatory T cells were detected in all six skin rash biopsies with a range of 0.2–5.5% cell proportion and were significantly lower in healthy skin (Fig. 6e ). In four out of six skin rash biopsies, IL-17A expression was noted in cells positive for panCK with a range of 2.2–14.6% cell proportion. In addition, colon biopsies collected through colonoscopy from five patients with immune-related colitis were compared to normal intestine tissue obtained from healthy individuals during routine colonoscopies (Fig. 6f–h ). Similar findings—that is, an increase of T H 17 and regulatory T cells as well as IL-17A expression—were noted in colon biopsies from patients with ir-colitis compared to normal intestine (Fig. 6i ). Specifically, there was a higher expression of CD4 + and IL-17A + cells compared to the normal intestine, with a range of 5.8–20.5% in ir-colitis samples and 0.3–0.7% in normal intestine tissue (Fig. 6j ). The abundance of regulatory T cells was increased in patients with ir-colitis, with a range of 3.9–17.2% cell proportion compared to 0.8–2.3% in normal intestine. Collectively, these observations complement the findings in the peripheral blood and show that CD4 + cells with IL-17A expression are upregulated at the site of the irAE occurrence.

a , H&E staining of a lichenoid skin rash biopsy. b , Six-plex immunofluorescence staining for IL-17A, FoxP3, CD4, CD8, CD68 and panCK of a lichenoid skin rash biopsy. c , Digital analysis of the multiplex immunofluorescence with cell annotation. T reg , regulatory T cell. d , Cell proportions of the lichenoid skin rash samples ( n = 6) compared to normal skin ( n = 4). e , Center log ratio normalized proportions of the lichenoid skin rash samples ( n = 6) compared to normal skin ( n = 4). Abundance means were compared using a two-sided Wilcoxon rank sum test and P values were not adjusted. f , H&E staining of an intestinal biopsy in a patient with ir-colitis. g , Six-plex immunofluorescence of an intestinal biopsy in a patient with ir-colitis. h , Digital analysis of the multiplex immunofluorescence with cell annotation. i , Cell proportions of the ir-colitis samples ( n = 5) compared to normal intestine ( n = 4). j , Center log ratio normalized proportions of the ir-colitis samples ( n = 5) compared to normal intestine ( n = 4). Abundance means were compared using a two-sided Wilcoxon rank sum test and P values were not adjusted.
Spatial transcriptomic analysis in inflamed tissue
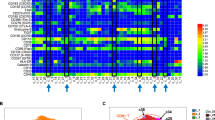
To further determine the transcriptomic profile of the cells involved at the site of the irAEs occurrence, we applied a spatial transcriptomic technology on the aforementioned samples of ir-colitis and lichenoid skin rash. Gene expression was measured in FFPE hematoxylin and eosin (H&E)-stained skin and colon sections using Visium technology (10x Genomics). The tissue sections were then spatially resolved in areas (spots) that were equally distributed over the tissue. The generated analysis of the transcriptomes of 4,306 spots and unsupervised clustering by UMAP revealed 11 major cellular clusters that were shared in the inflamed and normal intestine and skin tissue (Fig. 7a ). Clusters were defined by their marker gene expression and their spatial location on the analyzed tissue (Fig. 7b ). Genes encoding B cells and T cells, as well as T cell checkpoint molecules and macrophages were detected in the different cell types (Fig. 7c ). Of note, there were clusters with mixed cell types, such as fibroblasts and epithelial cells, with expression of B cell and T cell markers (CD19, CD8A, IFNγ, LAG3) as well as markers from the macrophage lineage, which were identified as ‘fibroblasts and immune cells’ and‘ epithelial and immune cells’, respectively. Proportion analysis of the identified transcriptomic subpopulations in six lichenoid skin rash biopsies compared to normal skin showed an increase in fibroblasts and immune cells (range, 1–19.5% cell proportion) and T cells and B cells (range, 0.5–40.4% cell proportion) (Fig. 7d ). Similar findings were noted in the four colon biopsies of patients with ir-colitis compared to normal intestine; there was an overall increase of epithelial and immune cells (range, 19.9–33.3% versus 0.5% cell proportion), fibroblasts and immune cells (range, 2.7–28.1% versus 0.3% cell proportion) and T cells and B cells (range, 0.3–5.9% versus 0–0.6% cell proportion) (Fig. 7d ). The expression of the major identified cell types, as well as the major type I and III cytokines, namely IFNG, IL6 and IL17A , was then examined in spatial resolution in the H&E samples (Fig. 8a–p ). This analysis showed an enrichment of the main type I and III cytokine transcripts, mainly in the upper dermis layers of the lichenoid skin rash. The respective cytokine transcripts in ir-colitis samples were also unequally distributed across the samples and were significantly enriched in areas of fibroblasts and immune cells, epithelial and immune cells, as well as T cells and B cells. A Wilcoxon rank sum test analysis for the pathway enrichment scores of type I and III cytokine transcripts showed significant differences between normal and inflamed biopsy specimen tissue (all, P < 0.01) (Fig. 8q–s ). Taken together, these results confirm that cytokine transcripts of type I and III immune responses are present at the site of the irAE occurrence.

a , UMAP plot with overview of the main cell types of the transcriptomes in the analysis of normal ( n = 2) and inflamed intestine ( n = 4), as well as normal ( n = 2) and inflamed skin tissue ( n = 6). b , Heatmap of the identified clusters and their marker gene expression. c , Dot plot with gene cell markers, including B cells and T cells ( CD19 and CD8A , respectively), macrophages ( CD68 ), keratinocytes ( KRT10 ), epithelial cells ( PIGR ), fibroblasts ( COL1A1 ), myofibroblasts ( MYH11 ) and apoptotic cells ( MT-ND5 ). d , Proportion analysis of the identified transcriptomic subpopulations in inflamed and normal tissue.
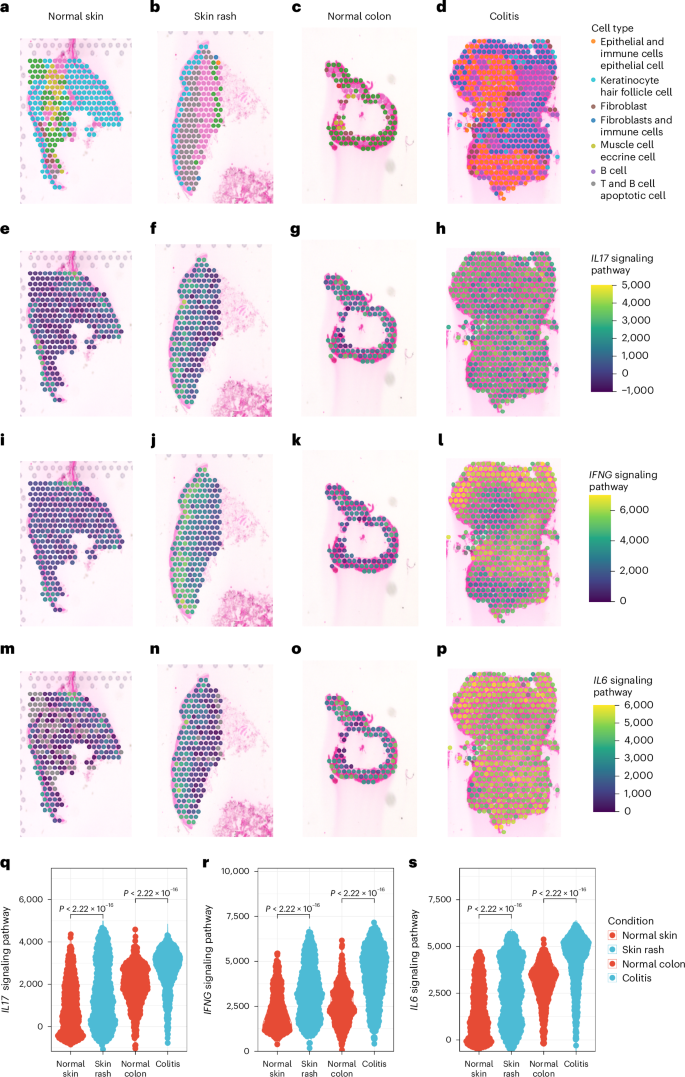
a – p , Representative spatial transcriptomic sections for normal skin ( n = 2) ( a , e , i , m ), lichenoid skin rash ( n = 6) ( b , f , j , n ), normal colon ( n = 2) ( c , g , k , o ) and immune-related colitis samples ( n = 4) ( d , h , l , p ) with the major identified cell types and enrichment scores of signaling pathways of type I and type III immunity. q – s , Wilcoxon rank sum test analysis for the pathway enrichment scores of type I and III cytokine transcripts for the IL-17 signaling pathway ( q ), IFNγ signaling pathway ( r ) and IL-6 signaling pathway ( s ); each dot represents one spot. The statistical test used was two-sided and P values were adjusted by the Benjamini–Hochberg method.
IL-17A blockade for the treatment of severe and refractory irAEs
Based on these results indicating an increase in CD4 + T cells with IL-17A expression at the site of inflammation, as well as the significant increase of IL-17A from TP 0 to TP 1 in patients with irAEs in all study cohorts, and given the availability of drugs targeting IL-17A, we next reasoned that IL-17A blockade diminishes severe, treatment-refractory irAEs. Therefore, we performed a focused clinical validation in a proof-of-concept case study. Two patients with stage IV melanoma received an anti-IL-17A monoclonal antibody (mAb), secukinumab, for three indications that included lichenoid skin rash grade 3 (according to the Common Terminology Criteria for Adverse Events v.5.0 (CTCAE v.5)), mild immune-related myocarditis (with concomitant immune-related myositis) grade 2 and immune-related colitis grade 3 (Supplementary Table 2 ). Of note, these irAEs occurred in the context of multiple, concurrent irAEs and were refractory to previous corticosteroid (solumedrol; max. dose of 250 mg day −1 ) and immunomodulatory treatment (TNF inhibitor mAb; max. dose of 5 mg kg −1 once to twice), as none of these treatments led to resolution of the irAEs of ≤grade 1. Both patients were treated with ipilimumab and nivolumab. The mild immune-related myocarditis was diagnosed based on the typical time window after ICI treatment, recurrence of previous immune-related myocarditis upon ICI re‐challenge, elevations of creatine kinase, high‐sensitive troponin T (hs‐TnT) and I (hs‐TnI), mild NT‐proBNP increase and positive magnetic resonance imaging criteria. Diagnosis of this mild myocarditis was underlined by an asymptomatic course without electrocardiogram changes or hemodynamic instability. After anti-IL-17A treatment, symptom improvement (≤grade 1) and/or complete resolution of the irAEs was noted within 43 days for the lichenoid skin rash, 20 days for the mild ir-myocarditis and 40 days for the ir-colitis. IL-17A levels were measured at TP 0 as well as before and after the administration of anti-IL-17A. In the following analysis of available serum samples in these patients, IL-17A increased from TP 0 to the time point before anti-IL-17A administration and decreased after the anti-IL-17A administration in both patients (Extended Data Fig. 7 ). Secukinumab was administered subcutaneously at a dose of 300 mg weekly for weeks 0–4, followed by 300 mg every 4 weeks. The total number of doses administered was two injections for the treatment of the mild ir-myocarditis and ir-colitis, whereas the patient with the lichenoid skin rash required a 150-day treatment course until complete resolution and discontinuation of secukinumab. The latter did not show any flares or relapses while on treatment and the patient did not require any local corticosteroids. In both patients, treatment with ipilimumab and nivolumab was permanently discontinued. Notably, the first patient was re-challenged with single-agent anti-CTLA4, without flare of the mild ir-myocarditis after a single dose of prophylactic secukinumab given 2 days before anti-CTLA4. The best overall response for the first patient was progressive disease and for the second patient was partial response.
We conducted a translational study to unravel the immune biology of the irAEs induced by ICIs and to propose possible treatment strategies in addition to the standard use of corticosteroids. For that purpose, we designed a prospective study with a precise longitudinal clinical sampling and analysis of serum samples acquired at the time of the irAEs onset. We show aberrant T cell activity with differential expression of type I and III immune signatures in the peripheral serum at the onset of irAEs that differed from the baseline immune signatures. This increase in circulating cytokines was particularly noted in patients with severe (grade ≥3) irAEs and in those with multiple concurrent irAEs. The observed immune signature was in line with an increase in the proportion of CD4 + T cells with IL-17A expression at the irAEs onset in the peripheral blood using flow cytometry. In contrast to the peripheral blood, multiplex immunofluorescence analysis of human lichenoid skin rash and ir-colitis samples identified a higher than expected proportion of CD4 + T cells with IL-17A expression. Spatial transcriptomics analysis of the lichenoid skin rash and ir-colitis samples confirmed the notion that type I and III immune responses are involved at the site of the irAE occurrence.
Altogether, and alongside the existing literature evidence, we reason that type I and type III responses are interconnected and that pro-inflammatory T H 17 cells show double-positive T H 1/T H 17 features that are associated with the onset of irAEs in patients treated with ICIs 42 , 43 . This concept is further supported by the identification of cytokines and chemokines that are induced by IFNγ, such as CXCL9 in the peripheral serum. Similar to autoimmune diseases, IL-17-expressing and IFNγ-expressing T cells are preferentially found at sites of inflammation 44 , 45 , whereas after tissue invasion, IL-17-expressing T cells produce high levels of IFNγ, indicating a high degree of plasticity 16 , 43 . Capitalizing on these results, we considered that IL-17A blockade can be used as a therapeutic target to mitigate toxicity in patients with corticosteroid-refractory irAEs. Clinical validation of these results with the administration of anti-IL-17A blockade in two patients who were treated with ipilimumab and nivolumab and who showed a serum increase of IL-17A at the irAEs onset resulted in resolution of severe, treatment-refractory irAEs.
These findings suggest a central role of the type III cells as one of the pathophysiological mechanisms for the development of irAEs. These results are in line with those previously demonstrated in translational studies, showing that expression of IL-6, a T H 17-cell differentiation cytokine, increased in inflamed, ir-colitis tissue of patients and mice treated with ICIs 20 . In addition, IL-17-producing cells have been linked with the pathogenesis of anti-CTLA4-driven ir-colitis 46 , 47 , whereas a predominantly T H 1 phenotype has been identified in ir-colitis and ir-dermatitis tissue in patients treated with ICIs 48 . Nevertheless, the exact nature of the latter (psoriasiform, lichenoid, maculopapular) has not been accurately defined in the referred study. In accordance with these results, we can show an enrichment of the main type I and (predominantly) type III cytokine transcripts in the spatial analysis, which was prevalent in both lichenoid skin rash and ir-colitis tissue. Respectively, PD1 blockade has been found to shift the antigen-induced cellular reactivity to a T H 1/T H 17 response 49 . Notably, although IL-17 is predominantly produced by CD4 + T cells upon recognition of peptide antigens presented by major histocompatibility complex molecules, several other T cell populations, such as γδ T cells and natural killer T cells, have also been shown to produce IL-17, the levels of which are usually low in the peripheral blood 38 , 39 , 50 . The differentiation of these cells is also IL-6, TGF-β and STAT3-induced 51 . Given that IL-17 has multiple pro-inflammatory properties, it has been previously suggested that selective modulation of the T H 17 differentiation rather than blocking the IL-17 cytokine per se might be more efficient in the downregulation of this pathway 52 . Further characterization of the immune signature of these IL-17-secreting cells in the context of the ICI-induced irAEs is required to guide personalized treatment decisions in the clinic.
These data underline the association of candidate serum proteins with the development of severe irAEs and may help identify therapeutic targets for their management. It has been previously shown that 11 circulating cytokines, including pro-inflammatory cytokines such as IL-1a, IL-2 and IFNa2, could predict the onset of severe irAEs in patients with melanoma that are being treated with anti-PD1-based ICIs 53 . In the current study, CXCL9, and particularly IL-17A, were consistently upregulated and were associated with the onset of irAEs in the main and external validation cohorts (for IL-17A). It has been shown that CXCL9 is induced by IFNγ, a type I cytokine 16 , and that IFNγ and TNF regulate the expression of CXCL16, which is expressed by colonic myeloid cells in patients with ir-colitis 54 . Additionally, CXCL9, CXCL10 and IFNγ are essential in recruiting effector T cells into tumors and have a substantial role in antitumor activity 55 . Similarly, TNF is produced by T H 1 cells and activated macrophages 56 , 57 , and increased TNF expression has been found in irAEs cases 20 , 48 . Taking into consideration the cross-induction of these cytokines, it is suggested to target specific cytokine pathways rather than a single downstream cytokine. In contrast to these type I responses, type III responses are less targeted to distinct immune effector cells 16 . Targeting the IL-17 production could treat ICI-induced toxicity, although its role in antitumor immunity has not yet been clearly specified. A recent study suggested that T H 17 cytokines support clinical benefit in patients with melanoma being treated with ipilimumab and nivolumab, but not single-agent anti-PD1 (ref. 58 ). In pancreatic cancer, IL-17 potentiates immunosuppressive effects through the recruitment of tumor-associated neutrophils, and subsequently, higher expression of IL-17 correlates with poorer prognosis 59 . Altogether, there is an inconsistency between the association of IL-17 with the tumor prognosis in the existing literature, which underlines that T H 17 cells are pleiotropic and their correlation with the treatment response is not linear. Consequently, prospective, randomized studies are required to elucidate the role of anti-IL-17A in the antitumor activity and the control of the irAEs.
We recognize that the present study has limitations. First, the included patients received either single-agent anti-PD1 or dual therapy using ipilimumab and nivolumab. These ICI treatment modalities are associated with different frequencies and severity grades of irAEs. In a phase 3 clinical trial for combined treatment with ipilimumab and nivolumab in advanced melanoma compared to treatment with nivolumab or ipilimumab alone, the type of toxicities did not differ between the two treatment agents 4 . Similarly, in the proteomic analysis of the present study, the changes in log 2 (fold change) at TP 0 and TP 1 were more dependent on the presence (and severity) of the irAEs rather than the treatment type; this observation should be further elucidated in future translational studies. Secondly, a correlation of these findings with the treatment response to ICIs is not feasible, given that the analyzed samples were specifically collected at the time point of the irAEs onset, and the median time from treatment initiation to serum collection was longer in patients with irAEs that in those without irAEs (37 days versus 22 days for the main and the internal validation cohorts). In addition, the heterogeneity of the tumor stages, treatment types, timing and type of investigations as well as the concurrent administration of corticosteroids and other immunosuppressive agents are not yet standardized and remain challenges in this analysis. Even though a positive correlation between tumor response to ICIs and increased risk for development of irAEs has been observed in randomized clinical and translational studies 60 , 61 , it remains unclear whether the timing of the onset of irAEs coincides with the response onset. We additionally recognize that some of the investigated proteins increased over time in patients without irAEs, and although these changes were not significant in the present study, this analysis is limited by the smaller sample size of these patients. Furthermore, in contrast to previous reports 53 , 62 , this study was not designed to identify predictive biomarkers for stratification of patients at risk for the development of irAEs, although its findings may support future experimental studies to further investigate their predictive value. Moreover, cytokine release in peripheral blood is transient and therefore poorly captured by most immunological assays, including scRNA-seq 63 . Similarly, T H 17 cells comprise only a small fraction of cells in the peripheral blood 64 . Lastly, the cytokine functions are pleiotropic and depend on the cell-type-specific receptor usage, such that characterization of these signaling activities at a single-cell level might be challenging 65 .
In conclusion, this study highlights the association of type III CD4 + T cells in the irAE development and provides proof-of-principle evidence to support a clinical trial examining anti-IL-17A for the management of irAEs. The differential expression of type I and III immune signatures in the peripheral serum at the irAEs onset, in particular in patients with severe (grade ≥3) irAEs or those with multiple concurrent irAEs early during the treatment course, emphasizes that these patients should be differentially classified and treated. The increase in the proportion of CD4 + T cells with IL-17A expression at the site of the inflammation suggests a key role of the type III responses across barrier tissues. Inhibiting these central inflammatory components may represent a more specific and effective therapeutic approach for the management of irAEs in addition to corticosteroid use. Correlative studies investigating the effect of anti-IL-17A in the antitumor response in a prospective, randomized setting will further highlight the multifunctional role of the T H 17 pathway in patients with melanoma treated with ICIs and will determine whether this approach is superior to other treatment strategies.
Ethics approval and consent to participate
Informed consent was obtained from all patients and healthy tissue donors, and all study analyses conformed to the principles set out in the World Medical Association Declaration of Helsinki and the Department of Health and Human Services Belmont Report. Healthy abdominal skin was obtained from consenting adult individuals and provided by the plastic surgery department of the University Hospital of Zurich with the assistance of the SKINTEGRITY.CH biobank. PBMCs and serum from patients were collected by the University Research Priority Program Cancer Biobank. The use of material for research purposes was approved by the Zurich Cantonal Ethic Commission (KEK-Nr. 2020-01148 and KEK-Nr. 2017-00688) and the ethics committee of Hannover Medical School (Nr. 8685_BO_K_2019). The study was conducted in accordance with the Declaration of Helsinki guidelines.
The anti-IL-17A treatment was approved and reimbursed by the Federal Office of Public Health through a specific allowance in the Swiss health codes (article 71a KVV, from 27 June 1995). More information about Article 71a can be found on the website of the Swiss Department of Health ( https://www.bag.admin.ch/bag/en/home/versicherungen/krankenversicherung/krankenversicherung-leistungen-tarife/Arzneimittel/verguetung-arzneimittel-im-einzelfall.html ).
Study population and clinical samples
We included patients with advanced, stage III–IV (AJCC v.8) 66 melanoma treated with anti-PD1-based treatment in the Department of Dermatology at the University Hospital of Zurich. Patients received either nivolumab (240 mg every 2 weeks), pembrolizumab (200 mg every 3 weeks), or combination of nivolumab and ipilimumab (1 mg kg −1 nivolumab and 3 mg kg −1 ipilimumab every 3 weeks, followed by nivolumab monotherapy 240 mg every 2 weeks). Patients were treated in the metastatic setting. Previous treatments were allowed; however, irAEs from previous treatments had to be completely resolved or be ≤grade 1 at the time of the treatment initiation for inclusion in the study. Overall, previous ongoing irAEs were noted in four (5%) patients of the main study cohort and included immunotherapy-induced hypothyroidism as a result of previous immune-related thyroiditis in two patients, immune-related uveitis in one patient and immune-related hepatitis in one patient, with a flare of the immune-related hepatitis upon ICI re-challenge. The discovery cohort was chosen from a retrospective cohort of 53 patients treated between January 2017 and August 2020, with available blood samples at baseline and the onset of the irAE. For the final analysis of the discovery cohort, nine patients with severe (≥grade 3, CTCAE v.5) irAEs that occurred early during the treatment and with available blood samples at baseline (0–29 days before therapy initiation) and at the irAEs onset were chosen. The discovery cohort was used as an exploratory cohort for the correlation of specific cytokines and chemokines with the irAEs onset.
The main study cohort was derived after the establishment of a study protocol with a prospective sample collection in patients treated with anti-PD1-based ICIs, including both single-agent anti-PD1 (pembrolizumab and nivolumab) and anti-PD1 in combination with anti-CTLA4 (ipilimumab). In this prospective study cohort, blood samples were prospectively collected at baseline (0–29 days before therapy initiation) and at regular intervals during therapy from August 2019 to March 2021. For the final analysis of the prospective study cohort, blood samples at baseline (0–29 days before therapy initiation) and at the irAEs onset or at the second to fifth infusion (in patients without any irAEs) were chosen.
An independent external validation cohort of patients with advanced, stage III–IV (AJCC v.8) melanoma started ICI treatment from October 2019 to February 2022 in the Department of Dermatology at Medical School Hannover was used to validate these results. Patients received either anti-PD1-based monotherapy (nivolumab) or a combined treatment with nivolumab and anti-CTLA-4 (ipilimumab). Blood samples taken at baseline and at irAEs onset or at the second to fifth infusion (in patients without any irAEs) were chosen. All serum samples were separated at 1,500 g for 10 min and the extracted serum was stored at −80 °C until further analysis. Patient demographics and clinicopathologic features included sex, age, disease stage (AJCC v.8), baseline lactate dehydrogenase (LDH) and Eastern Cooperative Oncology Group (ECOG) performance status for prior therapy were collected. Treatment characteristics, including investigator-assessed treatment response, were assessed radiologically at regular time intervals and according to the RECIST 1.1 criteria. Patients who achieved complete or partial response or stable disease were defined as ‘responders ’. Toxicity data were assessed using CTCAE v.5. Follow-up duration was calculated from the date of treatment initiation to the date of death, loss of follow-up or December 2022 (data cutoff).
Patient demographics and clinicopathologic features were collected for all patients included in the study. These included sex, age, disease stage (AJCC v.8), baseline LDH, previous treatments as well as the presence of autoimmune disease and ongoing toxicities from previous treatments at the treatment initiation. Treatment characteristics, including investigator-assessed treatment response, were assessed radiologically at regular time intervals and according to the RECIST 1.1 criteria 67 . Patients who achieved complete or partial response or stable disease for >6 months after treatment initiation were defined as ‘responders’. The best overall response was evaluated per the last available follow-up. Toxicity data were assessed using CTCAE v.5. Follow-up duration was calculated from the date of the treatment initiation to the date of death, loss of follow-up or August 2022 (data cutoff).
Blood samples
Blood samples, including serum and PBMCs, were collected at baseline (0–29 days before therapy initiation) and at regular time points during the therapy, until cycle 4. In the case of an irAE after cycle 4, additional blood samples were collected at irAE onset.
Multiplex cytokine–chemokine assay
Undiluted serum samples were profiled using the U-PLEX Assay Platform (MSD), as previously described 68 . In brief, biotinylated capture antibodies are coupled to U-PLEX linkers, which self-assemble onto unique spots on the U-PLEX plate. Analytes in the sample bind to the capture reagents; detection antibodies conjugated with electrochemiluminescent labels (MSD GOLD SULFO-TAG) bind to the analytes to complete the sandwich immunoassay. Once the sandwich immunoassay is complete, the U-PLEX plate is loaded into an MSD instrument, where a voltage applied to the plate electrodes causes the captured labels to emit light. The instrument measures the intensity of emitted light (which is proportional to the amount of analyte present in the sample) and provides a quantitative measure of each analyte in the sample. Each run was performed in duplicate for the discovery cohort; duplicates did not vary by more than 4%. A customized U-PLEX assay for the analysis of the following cytokines and chemokines was used in the discovery cohort: eotaxin, fractaline (CX3CL1), G-CSF, GM-CSF, IFNα2a, IFNβ, IFNγ, IL-1α, IL-1β, IL-2, IL-3, IL-4, IL-5, IL-6, IL-8, IL-9, IL-10, IL-12p70, IL-13, IL-17A, IL-18, IL-21, IL-22, IL-23, IP-10, I-TAC, MCP-1, MIP-1α, MIP-1β, TNF, TNFβ (LTA), VEGF-A and VEGF-D. For the main study cohort, the same assay without IL-3, IL-9, IL-13 and VEGF-D was used. For the external cohort, a V-Plex assay according to the manufacturer's instructions was applied. Data were collected by the Discovery Workbench v.4.0 (MSD).
Olink proteomics
Proteome analysis of baseline and on-treatment serum samples from the main study and internal validation cohort has been carried out using the Olink Explore 384 Inflammation panel (Olink Proteomics). Data collection was performed by Olink. This panel provides a high-throughput, multiplex immunoassay enabling analysis of 384 inflammation-related protein biomarkers across 96 samples simultaneously, using the proximity extension assay technology, as previously described 69 . In brief, pairs of oligonucleotide-labeled antibody probes bind to their targeted protein, and if the two probes are brought in proximity, the oligonucleotides will hybridize in a pair-wise manner. The addition of a DNA polymerase leads to a proximity-dependent DNA polymerization event, generating a unique target sequence analyzed through either next-generation sequencing or real-time PCR. Data were quality controlled and normalized using an internal extension control and an inter-plate control, to adjust for intra-run and inter-run variation. The final assay read-out is presented in NPX values, which is an arbitrary unit on a log 2 scale in which a high value corresponds to a higher protein expression. All assay validation data are available on the manufacturer’s website ( www.olink.com ).
Multiplex immunofluorescence
BOND RXm fully automated staining system was used in conjunction with the Opal Polaris 7-Color Automated IHC Detection Kit (NEL871001KT, Akoya Biosciences) according to user manual instructions. All antibodies were used at a dilution of 1:100 (IL-17A: ab79056, Abcam; FoxP3: 14-4777-82, Invitrogen, eBioscience; CD4: ab133616, Abcam; CD8: ab4055, Abcam; CD68: ab213363, Abcam; panCK: sc-8018, Santa Cruz Biotechnology). Slides were imaged simultaneously using the PerkinElmer Vectra Polaris imaging system maintained by the Center for Microscopy and Image Analysis, University of Zurich. Slide visualization and regions of interest selection were performed in Phenochart whole-slide viewer (Akoya Biosciences). Whole-slide scans can be made available on reasonable request. Cell segmentation training was performed according to DAPI nuclear staining, followed by cell marker phenotyping (CD4, CD8, CD68, FoxP3, IL-17A and panCK). Data were collected with InForm v.2.6.0 (Akoya Biosciences). In-depth spatial expression analysis was performed using Giotto (v.2.0.0.957) 70 .
scRNA-seq sample processing
Live cell biobanked PBMC samples were quickly thawed in a water bath set to 37 °C, re-suspended in 10 ml ice-cold RPMI (Sigma-Aldrich, cat. no. R0883) with 0.04% BSA (Sigma-Aldrich, cat. no. A7906) and incubated for 10 min on ice to allow the dimethylsulfoxide to diffuse from the tissue. Samples were spun down at 300 g for 5 min and re-suspended in PBS with 0.04% BSA (Sigma-Aldrich, cat. no. A7906); cell count and viability were assessed using Luna-FL cell counter (Dual Fluorescence Cell counter, Logos Biosystems, cat. no. L1001) using acridine orange propidium iodide (Logos Biosystems, cat. no. F23001) live/dead staining. Cell concentration was set according to 10x Genomics recommendation to 700–1,000 cells per μl; 10,000 cells were targeted per sample. Cell suspensions were loaded on Chip K (10x Genomics, PN-1000286) and processed using the 10x Genomics Chromium platform with the 5PV(D)J immune profiling kit v.2 on a 10x Genomics Chromium Single Cell Controller (10x Genomics, PN-120263). Gene expression libraries were amplified and sequenced on the Illumina NovaSeq 6000 platform at recommended sequencing depth (20,000–30,000 reads per cell for GEX libraries).
scRNA-seq analysis
Raw sequencing data were processed using the 10x Chromium Cell Ranger pipeline (v.7.0.0) ( https://support.10xgenomics.com/single-cell-gene-expression/software/downloads/latest ). Reads were aligned to the human reference genome (GRCh38, 2020-A) (10x Genomics). Single cells were filtered with the following parameters; >500 genes and <9,000 genes detected, <40% mitochondrial RNA and defined as ‘singlet’ by scDblFinder v.1.18.0 (ref. 71 ). The Seurat v.4 pipeline using SCTransform was used to normalize gene expression data 72 . Data integration was performed using 3,000 most variable features and the reciprocal PCA method using 30 dimensions. Dimension reduction was performed with PCA, followed by Leiden clustering using a resolution of 2.5. Visualization onto two-dimensional space was performed with UMAP. Cell typing was performed with SingleR 73 (v.1.4.1) using the Monaco Immune dataset ( GSE107011; PMID: 30726743). Differential expression was performed with the FindMarkers function from the Seurat v.4 package. ggplot2 (v.3.5.0) was used for visualizing aggregated data.
Cell stimulation and intracellular cytokine staining for flow cytometry
Live cell biobanked PBMC samples were quickly thawed in a water bath set to 37 °C, re-suspended in 10 ml of warm RPMI-1640 (Sigma-Aldrich, cat. no. R0883) supplemented with 5 nM l -glutamine (Gibco, Thermo Scientific, cat. no. 25030-024), 1 mM sodium pyruvate (Sigma-Aldrich, cat. no. S8636), 10% heat-inactivated fetal bovine serum (Biowest, cat. no. S181H) and 1% penicillin–streptomycin (Gibco, Thermo Scientific, cat. no. 15140-122). Cells were seeded in round-bottom 96-well plates at 1 × 10 6 cells per well (or less when not enough cells were available) and stimulated with Cell Stimulation Cocktail plus protein transport inhibitors (ThermoFisherScientific, cat. no. 00-4975-93) for 4 h at 37 °C with 5% CO 2 .
For flow cytometry staining, the cells were first stained with a fixable viability dye (Zombie Aqua, BioLegend, cat. no. 423101) according to the manufacturer’s instructions. Cells were washed with PBS + 2% FCS and stained with the following surface antibodies: CD3 APC (BioLegend, cat. no. 317318), CD4 Alexa Fluor 700 (BioLegend, cat. no. 344621), CD19 FITC (BioLegend, cat. no. 302205), CD8 Brilliant Violet 711 (BioLegend, cat. no. 344733), TCRγδ PE (BioLegend, cat. no. 331209) and HLA-DR Pacific Blue (BioLegend, cat. no. 307623). After surface staining, cells were washed with PBS + 2% FCS, and intracellular staining was performed using the Cyto-Fast Fix/Perm Buffer Set (BioLegend, cat. no. 426803) according to the manufacturer’s instructions and anti-human IL-17A PE-Cy7 (BioLegend, cat. no. 512315) or Mouse IgG1, κ Isotype Ctrl Antibody PE/Cy7 (BioLegend, cat. no. 400125) at the same concentration. Samples were acquired on an LSRFortessa Cell Analyzer (BD Biosciences), and data were analyzed with FlowJo (v.10.8.1).
Spatial transcriptomics Visium library preparation and sequencing
Sample processing and library preparation were performed according to the 10x Genomics manuals for Visium CytAssist Spatial Gene Expression for FFPE, using the human probe set v.2 and Visium slides with 6.5-mm capture areas. In short, FFPE samples were sectioned to 5-µm thickness and placed on Superfrost Plus slides. Sections were deparaffinized, H&E-stained and imaged on a Roche Ventana DP 200 Slide Scanner followed by destaining, decrosslinking and 16 h of probe hybridization. Ligated probes were transferred to Visium slides using the CytAssist instrument with standard parameters (37 °C and 30 min). Quality control was performed on the Roche LightCycler 96 for pre-amplified libraries and on the Agilent 4200 Tapestation for the final indexed libraries.
Spatial transcriptomics sequencing
The libraries were pooled according to estimated Visium slide coverage (Loupe Browser, 10x Genomics) and paired-end sequenced on an Illumina NovaSeq 6000 platform (Sequencing configuration: read 1, 28; i7-index, 10; i5-index, =10; read 2, 91) using an S1 flowcell (100 cycles).
Spatial Visium analysis
Raw sequencing data were processed using the 10x Chromium SpaceRanger pipeline (v.2.0.0) ( https://support.10xgenomics.com/spatial-gene-expression/software/downloads/latest ). Reads were aligned to the human reference genome (GRCh38, 2020-A) (10x Genomics). Spots with fewer than ten genes were filtered out. The Seurat v.4 pipeline for spatial analysis using SCTransform was used to normalize the data. Dimension reduction was performed with PCA, and clustering was performed with FindNeighbors and FindClusters using default parameters. Data integration was performed with Harmony 74 . Clusters were annotated manually from markers defined by FindAllmarkers. Gene set enrichment was performed using the package ‘escape’.
Statistical analysis
Clinical data were analyzed using gtsummary (v.1.7.2). For analysis of the clinical data, continuous variables (for example, age and follow-up time) were compared with a Student’s t -test or Wilcoxon rank test, as appropriate. Categorical variables (for example, sex, stage and LDH) were analyzed with a chi-squared test or Fisher’s exact test, as appropriate. All statistical tests used were two-sided. The Benjamini–Hochberg method was used for multiple testing correction. A P value of <0.05 was deemed statistically significant.
For the analysis of the multiplex cytokine–chemokine assay, MSD electrochemiluminescence data were imported into the MSD Discovery Workbench (v.4.0) analysis software for quantification. Quantified serum proteins were then imported into R (v.4.3.0) for statistical analysis. A linear mixed model was used to evaluate differences in serum proteins between the samples with and without adverse events in association with the time of the adverse event onset and the treatment response and between TP 1 and TP 0 . Differences in the log 2 (fold changes) between TP 1 (time of the irAEs onset of second to fifth infusion) and TP 0 (baseline) were evaluated with the Wilcoxon rank test and corrected for multiple testing with the Benjamini–Hochberg method. A P value of <0.05 was deemed statistically significant.
For the analysis of the proteomic data, Olink proteomic data were imported with the OlinkAnalyze R package (v.3.7.0). Differences in the log 2 (fold changes) between TP 1 and TP 0 were evaluated with the Wilcoxon rank test and corrected for multiple testing with the Benjamini–Hochberg method. A linear mixed model was used to evaluate differences in serum proteins between the samples with and without adverse events in association with the time of the adverse event onset and the treatment response and between TP 1 and TP 0 . The Benjamini–Hochberg method was used for multiple testing correction. A P value of <0.05 was deemed as statistically significant.
For the multiplex immunofluorescence and the spatial transcriptomic analysis, centered log ratios were compared using a two-sided Wilcoxon rank sum test and P values were adjusted with the Benjamini–Hochberg method. For flow cytometry, significant differences in the cell proportion analyses were determined by a one-sided Wilcoxon rank sum test.
Statistics and reproducibility
No statistical methods were used to pre-determine sample sizes, but our sample sizes are similar to those reported in previous publications 53 . No data were excluded from the analyses. This is an observational study and data collection was not randomized. Experiments involving Olink proteomic data, scRNA-seq, multiplex immunofluorescence and spatial transcriptomics were performed once. For the multiplex cytokine–chemokine assay (MSD), each run was performed in duplicate for the discovery cohort; duplicates did not vary by more than 4%. The flow cytometry experiment was performed in three replicates. All techniques and reagents used for the analyses of this study were previously optimized and validated. The investigators were not blinded to allocation during experiments and outcome assessment. Data collection and analysis were not performed blind to the conditions of the experiments. The data were formally tested for normality and equal variances and did not pass. Thus, non-parametric tests, such as the Wilcoxon rank-sum test, were used to compare groups. Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Subject details
Participant information on gender and age was self-reported. Information regarding gender, race and socioeconomic status was not collected. Patient information including gender and age are reported in Supplementary Table 1 . Consent has been obtained for reporting and sharing individual-level data.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The data supporting the findings of this study are available within the manuscript. The scRNA-seq and Visium-aligned 10x single-cell files that support the findings of this study are available at https://doi.org/10.5281/zenodo.10390377 . Source data for Figs. 2 , 3 and 5 , as well as Extended Data Figs. 1 , 2 , 3 , 6 and 7 have been provided as Source Data files. All other relevant de-identified raw data related to the current study are available from the corresponding authors upon reasonable academic request and will require the researcher to sign a data access agreement with the University Hospital of Zurich. Individual patient-identifiable data are not available owing to concerns with patient identification. Source data are provided with this paper.
Code availability
Code to generate the figures is available at the GitHub repository https://github.com/pcheng84/AE_analysis .
Postow, M. A., Callahan, M. K. & Wolchok, J. D. Immune checkpoint blockade in cancer therapy. J. Clin. Oncol. 33 , 1974–1982 (2015).
Article CAS PubMed PubMed Central Google Scholar
Postow, M. A., Sidlow, R. & Hellmann, M. D. Immune-related adverse events associated with immune checkpoint blockade. N. Engl. J. Med. 378 , 158–168 (2018).
Article CAS PubMed Google Scholar
Robert, C. et al. Pembrolizumab versus ipilimumab in advanced melanoma. N. Engl. J. Med. 372 , 2521–2532 (2015).
Larkin, J. et al. Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N. Engl. J. Med. 373 , 23–34 (2015).
Article PubMed PubMed Central Google Scholar
Haanen, J. et al. Management of toxicities from immunotherapy: ESMO clinical practice guideline for diagnosis, treatment and follow-up. Ann. Oncol. 33 , 1217–1238 (2022).
Schneider, B. J. et al. Management of immune-related adverse events in patients treated with immune checkpoint inhibitor therapy: ASCO guideline update. J. Clin. Oncol. 39 , 4073–4126 (2021).
Ghisoni, E. et al. Late-onset and long-lasting immune-related adverse events from immune checkpoint-inhibitors: an overlooked aspect in immunotherapy. Eur. J. Cancer 149 , 153–164 (2021).
Wang, D. Y. et al. Fatal toxic effects associated with immune checkpoint inhibitors: a systematic review and meta-analysis. JAMA Oncol . 4 , 1721–1728 (2018).
Johnson, D. B. et al. Fulminant myocarditis with combination immune checkpoint blockade. N. Engl. J. Med. 375 , 1749–1755 (2016).
Teulings, H. E. et al. Vitiligo-like depigmentation in patients with stage III–IV melanoma receiving immunotherapy and its association with survival: a systematic review and meta-analysis. J. Clin. Oncol. 33 , 773–781 (2015).
Axelrod, M. L. et al. T cells specific for α-myosin drive immunotherapy-related myocarditis. Nature 611 , 818–826 (2022).
Caturegli, P. et al. Hypophysitis secondary to cytotoxic T-lymphocyte-associated protein 4 blockade: insights into pathogenesis from an autopsy series. Am. J. Pathol. 186 , 3225–3235 (2016).
Iwama, S. et al. Pituitary expression of CTLA-4 mediates hypophysitis secondary to administration of CTLA-4 blocking antibody. Sci. Transl. Med. 6 , 230ra245 (2014).
Article Google Scholar
Sullivan, R. J. & Weber, J. S. Immune-related toxicities of checkpoint inhibitors: mechanisms and mitigation strategies. Nat. Rev. Drug Discov. 21 , 495–508 (2022).
Das, R. et al. Early B cell changes predict autoimmunity following combination immune checkpoint blockade. J. Clin. Invest. 128 , 715–720 (2018).
Tuzlak, S. et al. Repositioning T H cell polarization from single cytokines to complex help. Nat. Immunol. 22 , 1210–1217 (2021).
O’Connor, W. Jr. et al. A protective function for interleukin 17A in T cell-mediated intestinal inflammation. Nat. Immunol. 10 , 603–609 (2009).
Sonnenberg, G. F. et al. Pathological versus protective functions of IL-22 in airway inflammation are regulated by IL-17A. J. Exp. Med. 207 , 1293–1305 (2010).
Conti, H. R. et al. T H 17 cells and IL-17 receptor signaling are essential for mucosal host defense against oral candidiasis. J. Exp. Med. 206 , 299–311 (2009).
Hailemichael, Y. et al. Interleukin-6 blockade abrogates immunotherapy toxicity and promotes tumor immunity. Cancer Cell 40 , 509–523.e6 (2022).
Tarhini, A. A. et al. Baseline circulating IL-17 predicts toxicity while TGF-β1 and IL-10 are prognostic of relapse in ipilimumab neoadjuvant therapy of melanoma. J. Immunother. Cancer 3 , 39 (2015).
Dimitriou, F., Hogan, S., Menzies, A. M., Dummer, R. & Long, G. V. Interleukin-6 blockade for prophylaxis and management of immune-related adverse events in cancer immunotherapy. Eur. J. Cancer 157 , 214–224 (2021).
Stroud, C. R. et al. Tocilizumab for the management of immune mediated adverse events secondary to PD-1 blockade. J. Oncol. Pharm. Pract. 25 , 551–557 (2019).
Tokunaga, R. et al. CXCL9, CXCL10, CXCL11/CXCR3 axis for immune activation—a target for novel cancer therapy. Cancer Treat. Rev. 63 , 40–47 (2018).
Yao, Z. et al. Human IL-17: a novel cytokine derived from T cells. J. Immunol. 155 , 5483–5486 (1995).
Lodolce, J. P., Burkett, P. R., Koka, R. M., Boone, D. L. & Ma, A. Regulation of lymphoid homeostasis by interleukin-15. Cytokine Growth Factor Rev. 13 , 429–439 (2002).
Lamy, P. J. et al. Quantification and clinical relevance of gene amplification at chromosome 17q12-q21 in human epidermal growth factor receptor 2-amplified breast cancers. Breast Cancer Res. 13 , R15 (2011).
Kontos, F. et al. B7-H3: an attractive target for antibody-based immunotherapy. Clin. Cancer Res. 27 , 1227–1235 (2021).
Boyman, O. & Sprent, J. The role of interleukin-2 during homeostasis and activation of the immune system. Nat. Rev. Immunol. 12 , 180–190 (2012).
Borst, J., Hendriks, J. & Xiao, Y. CD27 and CD70 in T cell and B cell activation. Curr. Opin. Immunol. 17 , 275–281 (2005).
Hayette, S. et al. FLRG (follistatin-related gene), a new target of chromosomal rearrangement in malignant blood disorders. Oncogene 16 , 2949–2954 (1998).
Karsunky, H., Merad, M., Cozzio, A., Weissman, I. L. & Manz, M. G. Flt3 ligand regulates dendritic cell development from Flt3 + lymphoid and myeloid-committed progenitors to Flt3 + dendritic cells in vivo. J. Exp. Med. 198 , 305–313 (2003).
Al-Obaide, M. A., Alobydi, H., Abdelsalam, A. G., Zhang, R. & Srivenugopal, K. S. Multifaceted roles of 5′-regulatory region of the cancer associated gene B4GALT1 and its comparison with the gene family. Int. J. Oncol. 47 , 1393–1404 (2015).
Maglione, D. et al. Two alternative mRNAs coding for the angiogenic factor, placenta growth factor (PlGF), are transcribed from a single gene of chromosome 14. Oncogene 8 , 925–931 (1993).
CAS PubMed Google Scholar
Platchek, M., Lu, Q., Tran, H. & Xie, W. Comparative analysis of multiple immunoassays for cytokine profiling in drug discovery. SLAS Discov. 25 , 1197–1213 (2020).
Zweig, M. H. & Campbell, G. Receiver-operating characteristic (ROC) plots: a fundamental evaluation tool in clinical medicine. Clin. Chem. 39 , 561–577 (1993).
Cui, W., Liu, Y., Weinstein, J. S., Craft, J. & Kaech, S. M. An interleukin-21–interleukin-10–STAT3 pathway is critical for functional maturation of memory CD8 + T cells. Immunity 35 , 792–805 (2011).
Akitsu, A. & Iwakura, Y. Interleukin-17-producing γẟ T (γẟ17) cells in inflammatory diseases. Immunology 155 , 418–426 (2018).
Veldhoen, M. Interleukin 17 is a chief orchestrator of immunity. Nat. Immunol. 18 , 612–621 (2017).
Apalla, Z. et al. European recommendations for management of immune checkpoint inhibitors-derived dermatologic adverse events. The EADV task force ‘Dermatology for cancer patients’ position statement. J. Eur. Acad. Dermatol. Venereol. 36 , 332–350 (2022).
Larkin, J. et al. Five-year survival with combined nivolumab and ipilimumab in advanced melanoma. N. Engl. J. Med. 381 , 1535–1546 (2019).
Schnell, A. et al. Stem-like intestinal Th17 cells give rise to pathogenic effector T cells during autoimmunity. Cell 184 , 6281–6298.e23 (2021).
Hirota, K. et al. Fate mapping of IL-17-producing T cells in inflammatory responses. Nat. Immunol. 12 , 255–263 (2011).
Nistala, K. et al. Th17 plasticity in human autoimmune arthritis is driven by the inflammatory environment. Proc. Natl Acad. Sci. USA 107 , 14751–14756 (2010).
Paroni, M. et al. Recognition of viral and self-antigens by T H 1 and T H 1/T H 17 central memory cells in patients with multiple sclerosis reveals distinct roles in immune surveillance and relapses. J. Allergy Clin. Immunol. 140 , 797–808 (2017).
Bamias, G. et al. Immunological characteristics of colitis associated with anti-CTLA-4 antibody therapy. Cancer Invest. 35 , 443–455 (2017).
Callahan, M. K. et al. Evaluation of serum IL-17 levels during ipilimumab therapy: correlation with colitis. J. Clin. Oncol. 29 , 2505–2505 (2011).
Reschke, R. et al. Checkpoint blockade-induced dermatitis and colitis are dominated by tissue-resident memory T cells and Th1/Tc1 cytokines. Cancer Immunol. Res. 10 , 1167–1174 (2022).
Dulos, J. et al. PD-1 blockade augments Th1 and Th17 and suppresses Th2 responses in peripheral blood from patients with prostate and advanced melanoma cancer. J. Immunother. 35 , 169–178 (2012).
Papotto, P. H., Ribot, J. C. & Silva-Santos, B. IL-17 + γẟ T cells as kick-starters of inflammation. Nat. Immunol. 18 , 604–611 (2017).
Huber, M. et al. A Th17-like developmental process leads to CD8 + Tc17 cells with reduced cytotoxic activity. Eur. J. Immunol. 39 , 1716–1725 (2009).
Globig, A. M. et al. High-dimensional profiling reveals Tc17 cell enrichment in active Crohn’s disease and identifies a potentially targetable signature. Nat. Commun. 13 , 3688 (2022).
Lim, S. Y. et al. Circulating cytokines predict immune-related toxicity in melanoma patients receiving anti-PD-1-based immunotherapy. Clin. Cancer Res. 25 , 1557–1563 (2019).
Luoma, A. M. et al. Molecular pathways of colon inflammation induced by cancer immunotherapy. Cell 182 , 655–671.e22 (2020).
Groom, J. R. & Luster, A. D. CXCR3 ligands: redundant, collaborative and antagonistic functions. Immunol. Cell Biol. 89 , 207–215 (2011).
Kisuya, J., Chemtai, A., Raballah, E., Keter, A. & Ouma, C. The diagnostic accuracy of Th1 (IFN-γ, TNF-α, and IL-2) and Th2 (IL-4, IL-6 and IL-10) cytokines response in AFB microscopy smear negative PTB- HIV co-infected patients. Sci. Rep. 9 , 2966 (2019).
Parameswaran, N. & Patial, S. Tumor necrosis factor-α signaling in macrophages. Crit. Rev. Eukaryot. Gene Expr. 20 , 87–103 (2010).
Varaljai, R. et al. Interleukin 17 signaling supports clinical benefit of dual CTLA-4 and PD-1 checkpoint inhibition in melanoma. Nat. Cancer 4 , 1292–1308 (2023).
Zhang, Y. et al. Interleukin-17-induced neutrophil extracellular traps mediate resistance to checkpoint blockade in pancreatic cancer. J. Exp. Med. 217 , e20190354 (2020).
Das, S. & Johnson, D. B. Immune-related adverse events and anti-tumor efficacy of immune checkpoint inhibitors. J. Immunother. Cancer 7 , 306 (2019).
Xing, P. et al. Incidence rates of immune-related adverse events and their correlation with response in advanced solid tumours treated with NIVO or NIVO+IPI: a systematic review and meta-analysis. J. Immunother. Cancer 7 , 341 (2019).
Nunez, N. G. et al. Immune signatures predict development of autoimmune toxicity in patients with cancer treated with immune checkpoint inhibitors. Med 4 , 113–129.e7 (2023).
Stenken, J. A. & Poschenrieder, A. J. Bioanalytical chemistry of cytokines—a review. Anal. Chim. Acta 853 , 95–115 (2015).
Chen, G. et al. Th17 cell frequency and IL-17A production in peripheral blood of patients with non-small-cell lung cancer. J. Int. Med. Res. 48 , 300060520925948 (2020).
Ozaki, K. & Leonard, W. J. Cytokine and cytokine receptor pleiotropy and redundancy. J. Biol. Chem. 277 , 29355–29358 (2002).
Gershenwald, J. E. et al. Melanoma staging: evidence-based changes in the American Joint Committee on Cancer eighth edition cancer staging manual. CA Cancer J. Clin. 67 , 472–492 (2017).
Eisenhauer, E. A. et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur. J. Cancer 45 , 228–247 (2009).
Bolton, J. et al. Multiplex serological assay for establishing serological profiles of polymorphic, closely related peptide antigens. MethodsX 8 , 101345 (2021).
Assarsson, E. et al. Homogenous 96-plex PEA immunoassay exhibiting high sensitivity, specificity, and excellent scalability. PLoS ONE 9 , e95192 (2014).
Dries, R. et al. Giotto: a toolbox for integrative analysis and visualization of spatial expression data. Genome Biol . 22 , 78 (2021).
Germain, P. L., Lun, A., Garcia Meixide, C., Macnair, W. & Robinson, M. D. Doublet identification in single-cell sequencing data using scDblFinder. F1000Res 10 , 979 (2021).
Article PubMed Google Scholar
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184 , 3573–3587.e29 (2021).
Aran, D. et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 20 , 163–172 (2019).
Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16 , 1289–1296 (2019).
Download references
Acknowledgements
F.D. is supported by the Swiss Academy of Medical Sciences (SAMS) and the Gottfried and Julia Bangerter–Rhyner Foundation (YTCR 22/19). P.F.C. is supported by the Fonds zur Förderung des akademischen Nachwuchses (FAN) University of Zurich Alumni fellowship. The Melanoma Biobank is supported by the University Research Priority Program in Translational Cancer Research (U-402-04-02) and the SKINTEGRITY.CH Consortium Biobank is supported by a grant from the Monique Doronville de la Cour Stiftung and the ETH Foundation. M.P.L. is supported by a grant from the Swiss National Science Foundation (CRSII5_183478 / 1). We acknowledge the biobank team of the Dermatology Department of the University Hospital of Zurich and especially M. Schmid for her technical support and detailed work. We thank A. Ghosh and A. Cabral de Gouvea from the Functional Genomics Center Zurich, University of Zurich. We thank G. Restivo for providing us with the normal skin samples that were stained with multiplex immunofluorescence. We thank M. Tusup and S. Hogan for their important inputs in the study design. We thank all patients and families who participated in the study.
Author information
These authors contributed equally: Mitchell P. Levesque, Reinhard Dummer.
Authors and Affiliations
Department of Dermatology, University Hospital of Zurich, Zurich, Switzerland
Florentia Dimitriou, Phil F. Cheng, Annalisa Saltari, Ramon Staeger, Veronika Haunerdinger, Federica Sella, Aizhan Tastanova, Mitchell P. Levesque & Reinhard Dummer
Faculty of Medicine, University of Zurich, Zurich, Switzerland
Florentia Dimitriou, Phil F. Cheng, Annalisa Saltari, Ramon Staeger, Veronika Haunerdinger, Federica Sella, Aizhan Tastanova, Daniela Mihic-Probst, Mitchell P. Levesque & Reinhard Dummer
Department of Oncology, Geneva University Hospital, Geneva, Switzerland
Phil F. Cheng & Olivier Michielin
Department of Dermatology, Johannes Wesling Medical Center, Ruhr University Bochum Campus Minden, Minden, Germany
Katrin Schaper-Gerhardt & Ralf Gutzmer
Department of Dermatology, Medical School Hannover, Hannover, Germany
Katrin Schaper-Gerhardt
Functional Genomics Center Zurich, University of Zurich/ETH Zurich, Zurich, Switzerland
Christian Urban
Institute for Pathology and Molecular Pathology, University Hospital Zurich, Zurich, Switzerland
Susanne Dettwiler & Daniela Mihic-Probst
Department of Cardiology, University Heart Center and Center for Experimental Cardiology (CTEC), University Hospital Zurich and University of Zurich, Zurich, Switzerland
Christian M. Matter
Melanoma Institute Australia, The University of Sydney, Sydney, New South Wales, Australia
Georgina V. Long
Department of Medical Oncology, Royal North Shore and Mater Hospitals, Sydney, New South Wales, Australia
Faculty of Medicine and Health, The University of Sydney, Sydney, New South Wales, Australia
Institute of Experimental Immunology, University of Zurich (UZH), Zurich, Switzerland
Burkhard Becher
You can also search for this author in PubMed Google Scholar
Contributions
F.D., M.P.L. and R.D. conceived the study. F.D., P.F.C., A.S., R.S., V.H., A.T., S.D., M.P.L., G.V.L. and B.B. designed the methodology. F.D., P.F.C., A.S., D.M.P., K.S.G., R.G., F.S. and C.U. performed the investigation. F.D. and P.F.C. were responsible for the quality control of data and algorithms. F.D., P.F.C. and A.S. analyzed and interpreted the data. P.F.C. conducted statistical analysis. F.D. acquired funding. F.D. wrote the manuscript. F.D., P.F.C., A.S., A.T., K.S., R.G., V.H., G.V.L., B.B., M.P.L., C.M.M. and O.M. edited the manuscript. All authors revised the manuscript and approved the submission. All authors had full access to all the data and the final responsibility to submit for publication.
Corresponding authors
Correspondence to Florentia Dimitriou or Reinhard Dummer .
Ethics declarations
Competing interests.
This research received financial support from the Iten-Kohaut Stiftung Foundation and the University Hospital of Zurich (USZF27070) (F.D.), as well as from the Young Talents in Clinical Research program of the SAMS and Gottfried and Julia Bangerter–Rhyner Foundation (YTCR 01/22) (F.D.). F.D. receives or has received honoraria and travel support from Merck Sharp & Dohme, Bristol–Myers Squibb, Pierre Fabre and Sun Pharma outside the submitted work. K.S.G. receives research funding from Kyowa Kirin and Almirall. O.M. has a consulting advisory role for Bristol–Myers Squibb and Pierre Fabre, receives research funding from Bristol–Myers Squibb, Roche, Pierre Fabre, Amgen and MSD along with travel and accommodation expenses from Bristol–Myers Squibb. R.G. reports honoraria from Bristol–Myers Squibb, Merck Sharp & Dohme, Roche/Genentech, Novartis, Merck Serono, Almirall Hermal, Amgen, Sun Pharma, Pierre Fabre, Sanofi/Regeneron and Immunocore; a consulting or advisory role for Bristol–Myers Squibb, Merck Sharp & Dohme, Roche/Genentech, Novartis, Almirall Hermal, 4SC, Amgen, Pierre Fabre, Merck Serono, Sun Pharma, Sanofi, and Immunocore; research funding from Novartis, Amgen, Merck Serono, Sun Pharma, Sanofi, Almirall Hermal and Kyowa Kirin; and travel and accommodations expenses from Pierre Fabre, Sun Pharma and Boehringer Ingelheim. C.M.M. has received research grants to the institution from Eli Lilly, AstraZeneca, Roche, Amgen, Novartis, Novo Nordisk and MSD, including speaker or consultant fees. G.V.L. is consultant advisor for Agenus, Amgen, Array Biopharma, AstraZeneca, Boehringer Ingelheim, Bristol–Myers Squibb, Evaxion, Hexal AG (Sandoz Company), Highlight Therapeutics, Innovent Biologics USA, Merck Sharpe & Dohme, Novartis, OncoSec, PHMR, Pierre Fabre, Provectus, Qbiotics and Regeneron. M.P.L. has received project-specific research funding from Roche, Novartis, Molecular Partners and Oncobit. R.D. has intermittent, project-focused consulting and/or advisory relationships with Novartis, Merck Sharp & Dhome, Bristol–Myers Squibb, Roche, Amgen, Takeda, Pierre Fabre, Sun Pharma, Sanofi, Catalym, Second Genome, Regeneron, Alligator, T3 Pharma, MaxiVAX SA, Pfizer and touchIME, all of which took place outside the submitted work. All other authors declare no competing interests.
Peer review
Peer review information.
Nature Cancer thanks Genevieve Boland, Zlatko Trajanoski and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended data fig. 1 discovery cohort..
Concentrations (pg/ml) of cytokines and chemokines from baseline to the grade ≥3 immune-related adverse event (AE) onset in the discovery cohort. Boxplot with each protein by timepoint of n = 9 biological replicates for each timepoint (TP0 and TP1). Timepoint medians were compared using a two-sided Wilcoxon rank sum test and p values adjusted by the Benjamini-Hochberg method ( a ). Heatmap of log 2 fold change of serum cytokines and chemokines from baseline to grade ≥3 irAE onset for each patient (n = 9) ( b ).
Extended Data Fig. 2 Differential protein analysis by Olink in patient subgroups in the main study cohort.
Volcano plot with the differential expressed proteins at baseline (TP0) and at the AE onset (in patients with AEs) or at the second to fifth infusion (in patients without AEs) (TP1) in patients with multiple and single AEs (a) , severe and non-severe AEs (b) and according to treatment response in the main study cohort (n = 73) (c) . Significant difference was determined by two-sided Wilcoxon rank sum test and p -values were adjusted by the Benjamini-Hochberg method. Linear mixed effects regression analysis of the 11 significant proteins for the two time points (TP0 and TP1) according to treatment response, in n = 36 responders and n = 37 non-responders. Data represented by estimated marginal means with 95% confidence intervals. P -values are adjusted using the tukey method (d) . The y axis represents the Olink assay value (NPX).
Extended Data Fig. 3 Overview of the multiplex chemokine/cytokine assay analysis in the main study cohort.
Heatmap of log 2 fold change of the serum cytokines and chemokines per patient in the multiplex MSD assay, split by type of AE (none, single AE and multiple AEs) in the main study cohort (n = 73).
Extended Data Fig. 4 Uniform Manifold Approximation and Projection (UMAP) with overview of the main immune cell types split by time point and condition.
Single cell RNA sequencing analysis from the peripheral blood mononuclear cells (PBMCs) samples at the baseline and at the irAEs onset from three patients with AEs and at the baseline and second to fifth infusion in three patients without any AEs.
Extended Data Fig. 5 Flow cytometry (FC) gating strategy.
FC was used to determine IL-17A expression. Live cells were identified based on viability dye and SSC-A, and doublets were excluded based on FSC-H vs. FSC-A. T cells were identified based on expression of CD3, and CD4 + T cells were defined as CD4 + CD8-. For gating of IL17A+ cells, the threshold was set based on an IgG control staining and applied uniformly to all samples in the dataset.
Extended Data Fig. 6 Flow cytometry (FC) analysis from the collected PBMCs in six patients with (n = 3) and without (n = 3) irAEs.
Uniform Manifold Approximation and Projection (UMAP) with overview of the main identified cell types; the FC analysis included antibodies targeting CD4+ and CD8 + T-cells, NK cells, B cells, monocytes, dendritic cells and γδ T-cells were included ( a ). UMAP for IL-17A-producing cells ( b ). Proportional analysis of the IL-17A-producing CD4 + T-cells for the two time points (TP0 and TP1) in six patients with (n = 3) and without (n = 3) irAEs. Significant difference was determined by two-sided Wilcoxon rank sum test ( c ).
Extended Data Fig. 7 IL-17A serum concentration levels at the baseline (TP0), before and after the administration of anti-IL17A in the two patients that were treated with secukinumab.
Secukinumab treatment was administered subcutaneously at a dose of 300 mg weekly for weeks 0 to 4, followed by 300 mg every 4 weeks.
Supplementary information
Reporting summary, supplementary table 1.
Supplementary Table 1. Baseline characteristics in the main and validation cohort. Supplementary Table 2. Baseline characteristics and treatment outcome of patients‘ treated with anti-IL-17A (secukinumab) for the management of severe irAEs.
Source Data Fig. 2
Source data for heatmap, volcano plot and linear mixed model.
Source Data Fig. 3
Source data for heatmaps (a and c) and volcano plot.
Source Data Fig. 4
Source data for Fig. 4c,n.

Source Data Fig. 5
Source data for Fig. 5c,d.
Source Data Fig. 6
Source data for proportion (d,i) and boxplots (e,j).
Source Data Extended Data Fig. 1
Source data for violin plots (a) and heatmap (b).
Source Data Extended Data Fig. 2
Source data for volcano plots (a,b,c) and linear mixed model.
Source Data Extended Data Fig. 3
Source data for heatmap.
Source Data Extended Data Fig. 6
Source data for the flow cytometry.
Source Data Extended Data Fig. 7
Source data for the line plot.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/ .
Reprints and permissions
About this article
Cite this article.
Dimitriou, F., Cheng, P.F., Saltari, A. et al. A targetable type III immune response with increase of IL-17A expressing CD4 + T cells is associated with immunotherapy-induced toxicity in melanoma. Nat Cancer (2024). https://doi.org/10.1038/s43018-024-00810-4
Download citation
Received : 01 December 2023
Accepted : 18 July 2024
Published : 29 August 2024
DOI : https://doi.org/10.1038/s43018-024-00810-4
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Sign up for the Nature Briefing: Cancer newsletter — what matters in cancer research, free to your inbox weekly.
The blue social bookmark and publication sharing system.
Log in with your username.
I've lost my password.
Log in with your OpenID-Provider.
- Other OpenID-Provider
A case study of multiple-treatments meta-analysis demonstrates that covariates should be considered.

Comments and Reviews show / hide
Cite this publication, more citation styles.
BibSonomy is offered by the Data Science Chair of the University of Würzburg, the Information Processing and Analytics Group of the Humboldt-Unversität zu Berlin, the KDE Group of the University of Kassel, and the L3S Research Center .
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- PMC11343254

This is a preprint.
Evolutionary measures show that recurrence of dcis is distinct from progression to breast cancer, angelo fortunato.
1. Arizona Cancer Evolution Center and Biodesign Center for Biocomputing, Security and Society, Arizona State University, 727 E. Tyler St., Tempe, AZ 85281, USA
2. School of Life Sciences, Arizona State University, 427 East Tyler Mall, Tempe, AZ 85287, USA
Diego Mallo
Luis cisneros, lorraine m. king.
3. Duke University School of Medicine, Durham, NC 27710, USA
4. Department of Medicine, Genetics, and Biomedical Data Science Stanford School of Medicine, Stanford, CA 94305
5. Stanford Cancer Institute, Stanford School of Medicine, Stanford, CA 94305
Christina Curtis
6. Chan Zuckerberg Biohub, San Francisco, CA
Marc D. Ryser
Joseph y. lo, allison hall, jeffrey r. marks, e. shelley hwang, carlo c. maley, associated data.
All the sequencing data used in this manuscript is publicly available. The cross-sectional WES data at SRA with IDs (SRP298346 and XX) and the longitudinal WGS and WES data at HTAN dbGaP’s study accession phs002371.v6.p1.
Progression from pre-cancers like ductal carcinoma in situ (DCIS) to invasive disease (cancer) is driven by somatic evolution and is altered by clinical interventions. We hypothesized that genetic and/or phenotypic intra-tumor heterogeneity would predict clinical outcomes for DCIS since it serves as the substrate for natural selection among cells. We profiled two samples from two geographically distinct foci from each DCIS in both cross-sectional (N = 119) and longitudinal cohorts (N = 224), with whole exome sequencing, low-pass whole genome sequencing, and a panel of immunohistochemical markers. In the longitudinal cohorts, the only statistically significant predictors of time to non-invasive DCIS recurrence were the combination of treatment (lumpectomy only vs mastectomy or lumpectomy with radiation, HR = 12.13, p = 0.003, Wald test with FDR correction), ER status (HR = 0.16 for ER+ compared to ER-, p = 0.0045), and divergence in SNVs between the two samples (HR = 1.33 per 10% divergence, p = 0.018). SNV divergence also distinguished between pure DCIS and DCIS synchronous with invasive disease in the cross-sectional cohort. In contrast, the only statistically significant predictors of time to progression to invasive disease were the combination of the width of the surgical margin (HR = 0.67 per mm, p = 0.043) and the number of mutations that were detectable at high allele frequencies (HR = 1.30 per 10 SNVs, p = 0.02). These results imply that recurrence with DCIS is a clinical and biological process different from invasive progression.
Introduction
The improvement of radiological techniques and preventive screening of breast cancer conducted on a large scale makes it possible to identify mammary gland neoplasms at an early stage of development, when they are still confined within the glandular ducts. This neoplasm is termed ductal carcinoma in situ (DCIS) ( 1 ). Estimates from several natural history studies of DCIS indicate that 20–30% will progress to invasive cancer without definitive surgical treatment ( 2 , 3 ), implying that as many as 70% of patients who have surgery for DCIS may not derive benefit.
The ability to recognize which pre-cancerous tumors are likely to progress to invasive cancer is of great importance because it would identify high-risk patients for surgical, pharmacological, and/or radiation treatment. In contrast, low-risk patients could be managed by watchful waiting, avoiding the unnecessary harms and side effects associated with these therapies ( 4 ). Furthermore, selecting patients most at risk would facilitate reallocating healthcare resources to those who would benefit most from treatment.
Evolutionary mechanisms drive tumor progression ( 5 ). The impairment of control mechanisms of genetic integrity ( 6 ) accelerates the accumulation of new genetic alterations in cancer cells ( 7 ). The combination of these alterations in an increasing number of clones represents a critical factor in tumor progression, as these clones constitute the substrate upon which selection acts ( 8 ). The identification of mutations and the level of genetic (and phenotypic) heterogeneity have been shown to be associated with the risk of tumor progression in other pre-cancers, like Barrett’s esophagus ( 9 – 11 ). The higher the number of mutations and the greater the intratumor genetic heterogeneity, the higher the risk of developing clones that are cancerous, metastatic, and treatment-resistant ( 12 – 16 ).
It is challenging to integrate the combined effect of many mutations and genetic alterations that act simultaneously in cancer cells ( 17 ). Investigating the number of mutations and the level of heterogeneity allows us to introduce a quantitative parameter independent of the functional consequences of specific combinations of mutations, serving as a surrogate measure of the degree of evolvability of the neoplastic cells ( 18 , 19 ).
Both genetic and phenotypic heterogeneity can be measured by comparing different regions of the same tumor, ideally through analysis of longitudinal cohorts with linked clinical outcomes. Such studies often necessitate analysis of archival formalin-fixed paraffin-embedded (FFPE) samples, which is challenging due to partial degradation of the DNA, FFPE-induced artifacts, which manifest as sequence alterations, and low yield of nucleic acids from a limited number of sections. We recently published a workflow that overcomes these challenges, enabling the assessment of measures of genetic divergence between regions of the same tumor ( 20 ). This work aimed to test the hypothesis that genetic and phenotypic heterogeneity within DCIS can predict the recurrence of DCIS and/or progression to invasive ductal carcinoma (IDC).
Materials and Methods
Experimental design.
We performed two observational studies ( Fig. 1 , Table 1 ) to study DCIS progression. In a cross-sectional study ( Fig. 1A ), we compared DCIS samples from patients with DCIS only ( Pure DCIS , n = 58) versus DCIS samples from patients with synchronous invasive ductal carcinoma ( Synchronous DCIS , n = 61). In a longitudinal case-control study ( Fig. 1B ), we collected samples from patients with primary DCIS who were treated and then followed until they were diagnosed with an IDC recurrence ( Progressors , n = 56), were diagnosed with a DCIS-only recurrence ( Recurrents , n = 69), or did not recur within their follow-up time ( Nonrecurrents , n = 99, minimum five years). We calculated the median follow-up time using the reverse Kaplan-Meier method ( 21 ). In both cohorts, we characterized the genotype and phenotype of two DCIS regions per patient. All samples came from different FFPE blocks or were separated by at least 8mm. For some progressors, we also obtained a subsequent IDC sample. The Institutional Review Board (IRB) of Duke University Medical Center approved this study, and a waiver of consent was obtained according to the approved protocol.

A : Cross-sectional study: Synchronous DCIS tumors are presumed to have evolved from pure DCIS that existed before the progression of the synchronous IDC. In patients with synchronous DCIS, only the DCIS component was sampled and assayed unless otherwise specified. B : Longitudinal case-control study: pure-DCIS samples from patients treated and followed up for at least five years or until they progress or recur. n: number of patients per cohort.
Patient Cohorts with WES data, lpWGS, or IHC data.
| Study | Cross-sectional | Longitudinal | |||
|---|---|---|---|---|---|
| Assays | WES/IHC | WES/lpWGS/IHC | |||
| Cohort | |||||
| Number of patients | 58 | 61 | 99 | 69 | 56 |
| Median [min, max] | 2010 [1998, 2015] | 2010.5 [2000, 2017] | 2009 [2000, 2014] | 2008 [1998, 2017] | 2006.5 [1999, 2016] |
| Median [min, max] | 57.2 [34.0, 74.9] | 57.9 [40.9, 74.0] | 55.0 [41.0, 75.0] | 54.9 [40.0, 76.0] | 50.6 [38.0, 73.0] |
| 1 | 1 [1.7%] | 1 [1.6%] | 2 [2.0%] | 6 [8.7%] | 4 [7.1%] |
| 2 | 23 [39.7%] | 22 [36.1%] | 38 [38.4%] | 29 [42.0%] | 17 [30.4%] |
| 3 | 34 [58.6%] | 38 [62.3%] | 59 [59.6%] | 34 [49.3%] | 35 [62.5%] |
| Median [min, max] | 3.3 [0.8, 12.4] | 4.0 [0.2, 10.8] | 2.5 [0.3, 10.2] | 1.7 [0.2, 10.0] | 1.85 [0.4, 14.0] |
| ER (+) | 43 [74.1%] | 40 [65.6%] | 65 [65.7%] | 42 [60.9%] | 27 [48.2%] |
| ER (–) | 8 [13.8%] | 21 [34.4%] | 25 [25.3%] | 17 [24.6%] | 10 [17.9%] |
| ER (unknown) | 7 [12.1%] | 0 | 9 [9.1%] | 10 [14.5%] | 19 [33.9%] |
| Lumpectomy (Lump.) | 5 [5.1%] | 17 [24.6%] | 15 [26.8%] | ||
| Lump. + Radiation (Rad.) | 60 [60.6%] | 44 [63.8%] | 20 [35.7%] | ||
| Mastectomy | 22 [37.9%] | 24 [39.3%] | 33 [33.3%] | 7 [10.1%] | 19 [33.9%] |
| Lump. (Unk. Rad.) | 36 [62.1%] | 37 [60.7%] | 1 [1.0%] | 1 [1.4%] | 2 [3.6%] |
| Median [min, max] | 98 [60, 228] | 37 [12, 196] | 55 [12, 176] | ||
| Ink on tumor | 0 | 0 | 0 | ||
| <2mm | 23 [23.2%] | 28 [40.6%] | 15 [26.8%] | ||
| At least 2mm | 35 [35.4%] | 22 [31.9%] | 20 [35.7%] | ||
| Clear, NA mm | 41 [41.4%] | 19 [27.5%] | 21 [37.5%] | ||
| White | 34 [58.6%] | 43 [70.5%] | 67 [67.7%] | 41 [59.4%] | 28 [50.0%] |
| Black | 22 [37.9%] | 15 [24.6%] | 22 [22.2%] | 21 [30.4%] | 23 [41.1%] |
| Other | 2 [3.4%] | 3 [4.9%] | 2 [2.0%] | 2 [2.9%] | 2 [3.6%] |
| Unknown | 0 | 0 | 8 [8.1%] | 5 [7.2%] | 3 [5.4%] |
Unk. = Unknow.
Clinical specimens
We classified breast tumors according to the World Health Organization (WHO) criteria ( 22 ). We graded the IDC and DCIS samples according to the Nottingham grading system ( 23 ) or recommendations from the Consensus Conference on DCIS classification ( 24 ), respectively.
All samples were obtained from formalin-fixed paraffin-embedded (FFPE) breast tissue blocks. Cases from the cross-sectional studies were obtained from the Duke Pathology archives. Cases from the longitudinal study were obtained from Translational Breast Cancer Research Consortium (TBCRC) sites, a national multi-center consortium of cancer centers that treat breast cancer patients. All cases underwent detailed pathology review (AH) for histologic features and case eligibility.
DNA extraction and sequencing
The DNA extraction, sequencing, and data processing protocol has been previously reported ( 20 ). For each neoplastic sample, we extracted the DNA from multiple serial archival FFPE tissue block sections after macro-dissecting the areas of interest. To estimate the germline sequence, we also extracted DNA from either distant benign breast tissue or a benign lymph node. The study pathologist confirmed the presence of ≥70% neoplastic cells in the microdissected areas of neoplastic samples and their absence from control samples.
After DNA extraction, hybrid capture was performed using two targeted panels (all exons of the 83 genes in the breast cancer gene panel and the human exome), and the multiplexed libraries were sequenced using either an Illumina HiSeq with 4-channel chemistry (cross-sectional study) or a NovaSeq 6000 machine with 2-channel chemistry (longitudinal study). After alignment to the Genome Reference Consortium Human Build 37 and marking duplicates, we obtained a mean de-duplicated depth of 115.9 ± 52.2 (SD). The resulting BAM files were the input data for our SNV calling and heterogeneity calculation pipeline. We discarded samples with less than 40% of the target covered at 40X. Sequencing was performed at the McDonnell Genome Institute at Washington University School of Medicine in St. Louis.
Additionally, we performed low-pass whole genome sequencing for the longitudinal study as previously described ( 25 ). The resulting BAM files were used as the input data for the CNA characterization pipeline.
SNV characterization
We used our previously reported software ITHE ( 20 ) to calculate by-patient SNV burden and divergence, leveraging the two neoplastic geographically distant samples and a control sample from the same patient. We recently developed, optimized, and validated this pipeline using 28 pairs of technical replicates (same extracted DNA, two aliquots were independently sequenced) of macrodissected FFPE DCIS samples similar to the specimens analyzed here. We used the filtering parameters we found optimal previously ( 20 ). ITHE was optimized for accurate divergence estimation and thus tries to maximize variant calling’s precision. We measured SNV divergence as the percentage of mutations detected in the union of the mutations from the two samples that are not shared by both samples. We required that the union set of mutations had at least five mutations to calculate divergence. SNV burden was calculated as the union of mutations in both samples. When comparing DCIS and IDC samples in the cross-sectional study, we report the mean of the two comparisons between one of the two DCIS samples and the IDC sample.
Functional analysis
We performed the functional enrichment analysis of genes that harbored non-synonymous SNV mutations with PANTHER ( 26 ) and DAVID ( 27 , 28 ). We corrected the fold enrichment p-values considering the false discovery rate (FDR).
CNA characterization
We followed our previously published protocols for low-pass WGS data processing and CNA calling ( 25 ). Briefly, we used Nextflow-base’s Sarek pipeline to align the lpWGS data to the GRCh38/hg38 reference genome, marked duplicates, and re-calibrated quality scores. We used the resulting alignments to call autosomal CNA variants using QDNAseq ( 29 ) on 50-kb genomic bins after filtering genomic regions and reads for mappability and QC content while estimating ploidy and purity. We corrected the log2 ratio for the latter. CNAs with ∣ c o r r e c t e d l o g 2 r a t i o ∣ > 0.3 were considered as altered and normal otherwise. To maximize the robustness of our statistics, we measured CNA burden per sample as the proportion of the genome that was altered (over the total genome considered) and CNA divergence per patient as the proportion of the altered genome that is not shared between the two samples over the altered genome per patient (i.e., C N A d i v e r g e n c e = A Δ B A ∪ B , with A and B defined as the set of altered genomic regions of each homonymous sample, and Δ the set symmetric difference operator).
Immunohistochemistry characterization
We chose a series of 15 candidate proteins ( Supplementary Table S1 ) representing several categories including essential breast cancer drivers (ER, PR, HER2), immune-related (FOXP3, CD68), resource and microenvironmental measures (GLUT1, CA9, CD31, FASN), myoepithelial and basement membrane (TP63, COL15A1) and progenitor or stem cell-related (ALDH1 and RANK) markers. Additional proteins included the proliferation marker KI-67, the adhesion marker phospho-FAK, and COX2 (PTGS2), which were previously described as being associated with DCIS progression. In the longitudinal study, these were reduced to ER and GLUT1 only ( Supplementary Tables S2 - 3 ), based on the results from the cross-sectional study and the paucity of samples. We measured stain intensity using detailed expert scoring. In most cases, the study pathologist used a scoring system that captures the distribution of intensities in an IHC profile, while for a smaller number of markers, it was binary ( Supplementary Table S1 ). The IHC profile was quantified as the percentage of the slide presenting different levels of increasing staining intensity: absence, low, medium, and high. Medium staining was deemed as approximately twice as intense as low staining and high staining three times as intense as low staining.
We evaluated the IHC at three different scales of comparison:
- The average intensity of immunofluorescence across samples for each patient, measuring the typical intensity of IHC signal per patient.
- The variance of the intensity between samples for each patient, measuring the variations of IHC signal between distant locations in each patient.
- The variance of intensity within samples, measuring the variations of IHC signal at short distances in each patient.
These three measures are quantified by the Mean of Intensity Score, the Earth Mover’s Distance, and the Cumulative Density Index. Briefly, the Intensity Score is the weighted sum of the IHC profile proportions normalized by the maximum possible staining, the Earth Mover’s Distance represents the minimum cost of turning one profile into another ( 30 ), and the Cumulative Density Index represents how close from a uniform distribution the observed profile is and ranges from 0 (all the profile weight in one of the extreme categories) to 1 (uniform profile). See a detailed description of these statistics in Supplementary Text 1 .
Statistical analysis
Cohort characterization.
For each study, we compared differences in the central tendency of genetic and phenotypic variables per patient between cohorts using the Kruskal-Wallis Rank Sum or the Mann-Whitney U tests for many or two cohorts, respectively. We followed the Kruskal-Wallis Rank Sum test with Dunn’s post-hoc test while controlling for multiple tests using the Holm-Šidák adjustment ( 31 ). Exceptionally, CNA divergence met the assumptions of a parametric test, and thus, we used an ANOVA followed by Tukey HSD post-hoc tests. In cases where we used multiple measurements per patient (CNA burden), we used a Mixed-effects ANOVA with different random effect intercepts per patient to account for data dependencies (on the square-root-transformed variable), followed by Tukey’s HSD on the estimated marginal means.
Distinguishing Pure DCIS from Synchronous DCIS
We performed variable selection among the phenotypic measurements with significant differences between cohorts using a Random Forest classification model ( 32 ) under the Gini impurity criterion to return the importance ranking of each feature given by their predictive power. We used the two top measurements to build a generalized linear logistic model. Similarly, we built a generalized linear logistic model with the genetic measurements that showed significant differences between cohorts and the combination of the three. Due to missing data, we compared the models under the Akaike information criterion (AIC) on the smallest dataset for all models ( 33 ).
Association with clinical outcomes
Using our longitudinal study, we determined whether genetic and phenotypic statistics were independently associated with the time to clinical outcome (non-invasive recurrence or progression) using Cox regression analyses after checking they met the proportional hazards assumption. Nonrecurrent patients were right-censored using their follow-up time, and progressors’ recurrence time was used as their time to clinical outcome. Recurrents were discarded when considering progression, and progressors were discarded when considering non-invasive recurrence. We also provide supplementary results in which the clinical outcomes are “any recurrence” and “progression without discarding recurrent patients.” In this case, recurrents were right-censored at the time of recurrence when considering progression, and otherwise, their recurrence time was used as their time to clinical outcome. We evaluated the statistical significance of Cox regressors using the Wald test. We used the proportional hazard regression model for one variable (SNV burden) to stratify patients into low and high SNV burden and plotted their event-free survival curves. We stratified using the risk relative to the patient with all variables (i.e., SNV burden here) set at the mean value (i.e., type = “risk”, reference = “sample”, in the predict.coxph function of the survival R package). We chose the threshold that maximized Youden’s J statistic ( 34 ) using the true outcomes. In all cases, we used the log-rank test to compare the survival trends of two or more groups.
We also integrated 18 clinical covariates ( Supplementary Table S4 ) with our eight genetic and phenotypic measurements to model time to non-invasive recurrence and time to progression. We performed variable selection using Cox LASSO and chose the regularization parameter that minimized the partial-likelihood deviance via 10-fold cross-validation. To reduce the stochasticity of the results, we performed this process 100 independent times per model and selected the variables that were selected in at least 90% of them. To reduce missingness, we performed mean imputation on the clinical covariates before variable selection. The selected variables were used to build the final Cox regression models using all patients with available (imputed) data for those variables. Alternatively, we selected patients with data for all covariates chosen without imputation. We used the final models to stratify patients as in the univariate proportional hazards regression above. In all cases, the model used to stratify patients and plot their event-free survival curves includes all the variables included in the forest plot. All variables were standardized to make hazard ratios (HRs) comparable, and thus, HRs are relative to a change of 1 standard deviation unless specified otherwise.
Data Availability
Reproducibility.
Scripts to reproduce most data pre-processing and statistical analysis can be found at https://github.com/adamallo/ManuscriptScripts_DCISRecurrenceVsProgression .
Study cohorts
We investigated DCIS progression to invasive cancer using two independent observational studies with different patients: a cross-sectional study and a longitudinal study ( Fig. 1 , Table 1 ). In the cross-sectional study ( Fig. 1A ), we compared DCIS samples from patients with DCIS only ( Pure DCIS , n = 58) versus DCIS samples from patients with synchronous DCIS with invasive ductal carcinoma ( Synchronous DCIS , n = 61). In the separate longitudinal study ( Fig. 1B ), we compared pure DCIS samples from patients who were treated and had long-term follow-up (median = 117 months, 95% CI [104, 132]). This cohort consisted of patients who progressed to IDC ( progressors ) (n = 56), patients who had a DCIS-only recurrence ( recurrents , n = 69), or patients who did not recur during the follow-up interval ( nonrecurrents , n = 99). In both studies, we characterized the genotype and phenotype of two formalin-fixed paraffin-embedded DCIS samples per patient, enabling measures of evolutionary divergence (see Methods ). We also obtained a single sample of their IDC recurrence for some progressors.
Cross-sectional study
Single nucleotide mutational burden.
Pure DCIS carried fewer SNVs per patient (mean 7.5 ± 10.6 standard deviation) than synchronous DCIS (10.4 ± 15.3), but this difference was not statistically significant ( Fig. 2A ).

Distribution of the number of SNVs per patient in the two cross-sectional cohorts A and the two lesion types (DCIS vs. IDC) present in the synchronous cohort B . Distribution of SNV genetic divergence (percentage of private mutations) per patient in the two cross-sectional cohorts C . We calculated divergence for tumors with at least five mutations in the union of the two samples, which explains the lower number of tumors per group. P-values shown if p ≤ 0.1, A, C : Mann-Whitney U, B : Paired-samples sign test. Interquartile range (vertical line) and median (point) in burgundy, N: number of patients.
The invasive component in synchronous DCIS patients showed a statistically significantly increased number of SNVs (18.1 ± 31.5, Fig. 2B ) compared with their DCIS counterpart (Paired-samples sign test, p = 0.04) largely due to four cases of IDC with a dramatic increase in mutation burden.
SNV Genetic Divergence
We measured the SNV genetic divergence as the percentage of mutations that are private to either sample per patient. Synchronous DCIS showed higher genetic divergence (21.5% ± 17.5%) than pure DCIS (10.8% ± 17.4%, Fig. 2C ) (Mann-Whitney U test, p = 0.009). Additionally, we also characterized the genetic divergence between the two synchronous components (i.e., DCIS vs. IDC in synchronous patients) (44.5% ± 29.0%), which is higher than the paired synchronous DCIS divergence ( Supplementary Fig. S1 , Paired-samples sign test, p = 0.002).
Phenotypic characterization
Synchronous DCIS samples presented higher levels of GLUT1 staining ( p = 0.004) and lower levels of CA9 staining ( p = 0.01) than pure DCIS samples ( Fig. 3A , pairwise Mann-Whitney U tests of mean intensity scores [MIS], unadjusted p-values); all other markers showed non-significant differences between groups. This result holds when one of the two DCIS samples per patient is used randomly instead of the MIS ( Supplementary Fig. S2 ).

Distribution of mean intensity scores (MIS) per patient (see Methods ) A , between-sample divergence ( B , Earth Mover’s Distance [EMD]) and within-sample divergence ( C , Cumulative Density Index [CDI]). A: for each patient and IHC marker, B and C: only markers with significant differences between cohorts (unadjusted p-values). Unadjusted pairwise Mann-Whitney U p-values shown if p ≤ 0.1. Interquartile range (vertical line) and median (point) in burgundy. N: number of patients.
Phenotypic Divergence
We characterized the between-sample phenotypic divergence for each marker using a distance between staining intensity profiles (Earth Mover’s Distance) and the within-sample divergence using a measure of staining intensity uniformity (Cumulative Density Index; see Supplementary Methods for detailed definition of these indices).
Multiple markers presented differences in between-sample divergence between pure DCIS and synchronous DCIS samples, with the latter showing increased divergence for GLUT1 ( p = 0.01), FOXP3 ( p = 0.01), and HER2 ( p = 0.04) staining, but decreased divergence of ER ( p = 0.01) staining ( Fig. 3B , Supplementary Fig. S3 , Pairwise Mann-Whitney U tests, unadjusted p-values). This reduction of ER phenotypic divergence in synchronous DCIS samples was replicated in the within-sample measures ( p = 0.01) and mimicked by CA9 ( p = 0.01) ( Fig. 3C , Supplementary Fig. S4 , Pairwise Mann-Whitney U tests, unadjusted p-values). A reduction in the phenotypic divergence for ER in synchronous DCIS samples indicates larger uniformity across and within samples, while the mean intensity of ER signal is not markedly different ( Fig. 3A ).
All eight significant phenotypic divergence features—MIS for GLUT1 and CA9 ( Fig. 3A ), EMD for GLUT1, FOXP3, ER and HER2 ( Fig. 3B ), and CDI for ER and CA9 ( Fig. 3C )—were combined in a mixed logistic regression to model the progression status of the samples, from which the most important features were selected according to their relative predictive power. A reduced logistic model including between-sample diversity (EMD) for GLUT1 and within-sample diversity (CID) for ER had statistically significant coefficients (GLUT1 EMD, p = 0.01; ER CDI, p = 0.01) and spanned 40 pure DCIS cases and 52 synchronous DCIS cases. Therefore, we selected these two IHC markers (GLUT1 and ER) as the targets for phenotypic divergence to be included in the longitudinal study.
Logistic regression showed that the only statistically significant genetic measurement (SNV divergence) was strongly associated with the cohort, with p = 0.0136, so it was also selected for evaluation in the longitudinal study.
Longitudinal Study: Associations with Recurrence and Progression
We used the cross-sectional cohort as a discovery cohort, using the synchronous DCIS as a proxy for high-risk DCIS likely to progress to IDC. Samples in our validation cohorts come from patients with pure DCIS with known outcomes ( nonrecurrent , recurred as DCIS, progressed to IDC) and were obtained before treatment ( Fig. 1B ). We sequenced the exomes of two regions of each index DCIS in the longitudinal cohorts, mirroring the methods for the cross-sectional cohorts, and also performed low-pass whole genome sequencing data for most samples.
Mutational burden
Primary DCIS tissue from nonrecurrent patients carried the fewest SNVs (13.4 ± 18.2), followed by that of recurrent patients (19.2 ± 26.4) and progressors (39.7 ± 46.2). These relationships between cohorts were mirrored by the CNA alteration burden ( nonrecurrents : 15.9% ± 15.0% genome altered, recurrents : 17.3% ± 14.8%, progressors : 24.6% ± 17.1%) but presented higher p-values. Thus, SNV burden shows statistically significant differences between nonrecurrents and progressors ( p = 0.003) and between recurrents and progressors ( p = 0.05, Dunn’s test corrected for multiple tests with the Holm-Šidák adjustment) ( Fig. 4A ). In contrast, CNA burden was significantly different only between nonrecurrents and progressors ( p = 0.03, Tukey HSD) ( Fig. 4B ).

Distribution of SNV ( A , C ) and CNA ( B , D ) mutational burdens ( A , B ) and divergences ( C , D ) in the three longitudinal cohorts (Nonrec: nonrecurrents , Rec: recurrents , Prog: progressors ). A : number of SNVs per patient; Omnibus test: Kruskal-Wallis Rank Sum, Post-hoc test: Dunn’s test with control for multiple tests using the Holm-Šidák adjustment. B : proportion of genome with copy number alterations per sample; Omnibus test: Mixed-effects ANOVA on the square-root-transformed proportion of genome altered, Post-hoc test: Tukey HSD on estimated marginal means. C : percentage of private SNV mutations per patient; Omnibus test: Kruskal-Wallis Rank Sum. D : percentage of the genome with copy number alterations private to either sample per patient; Omnibus test: ANOVA, Post-hoc test: Tukey HSD. P-values shown if adjusted p ≤ 0.1. Interquartile range (vertical line) and median (point) in burgundy, N: number of data points ( A , C , and D : patients, B : samples). We only calculated divergence for tumors with at least five mutations in the union of the two samples, which explains the lower number of tumors in C .
Genetic Divergence
Similar to SNV divergence, we measured CNA divergence as the percentage of the altered genome that is private to either sample per patient. SNV divergence was highest in recurrent patients but not statistically different between cohorts ( nonrecurrents : 17.0% ± 13.8%, recurrents : 28.2% ± 25.5%, progressors : 18.4% ± 19.3%, Fig. 4C ). In contrast, CNA divergence followed a decreasing pattern of divergence with progression ( Fig. 4D ), by which nonrecurrents were the most divergent (77.4% ± 16.4%), followed by recurrents (67.7% ± 23.4%) and progressors (63.7% ± 21.7%). Only progressors and nonrecurrents showed statistically significant differences in CNA divergence in pairwise comparisons ( Fig. 7B , p = 0.03, Tukey HSD).

Forest plots describing proportional hazard regressions using variables selected with LASSO ( A , C ) and corresponding Kaplan-Meier plots of patients stratified by the relative risk threshold that maximizes Youden’s J statistic of the outcomes ( B , D ). A-B : Non-invasive-recurrence-free survival. C-D : Progression-free survival. Hazard Ratios (second column, A , C ) are relative to 1 standard deviation. Lumpectomy Only is compared to Lumpectomy + Radiation and Mastectomy and ER+ is compared to ER-. No microcalc(ification)s is compared to having microcalcifications in DCIS-only and/or benign ducts. Tables below Kaplan-Meier plots show the number of samples at risk at different times. Log-rank test.
Functional analysis of non-synonymous SNV mutations
The functional analyses highlighted significant differences between the three cohorts. According to DAVID , recurrent patients showed enrichment of mutated genes involved in taste reception (TAS2R30, TAS2R31, TAS2R43, and TAS2R46), while progressors showed enrichment of genes typically mutated in cancers such as endometrial, small cell lung, prostate, and breast cancer, glioma and melanoma (PIK3CA, ERBB2, PTEN, AKT1, PIK3R2, TP53, PIK3CG), and genes involved in the determination of cell shape, arrangement of transmembrane proteins, and organization of organelles (SPTA1, SPTBN5, DST, SPTAN1). Nonrecurrents did not show significant functional enrichment ( Supplementary Table S5 ). In addition, PANTHER functional analysis revealed an enrichment of several pathways only in progressors ( Supplementary Table S6 ), such as Hypoxia response via HIF activation ( p < 0.001, false discovery rate correction herein this section), Insulin/IGF pathway-protein kinase B signaling cascade ( p < 0.001), p53 pathway ( p = 0.003), Endothelin signaling pathway ( p = 0.003), Hedgehog signaling pathway ( p = 0.02), and PI3 kinase pathway ( p = 0.03).
Phenotypic Characterization and Divergence
We characterized the DCIS phenotypes of the three cohorts using the immunohistochemical profiles of the two markers that showed the highest discriminating power between the two cross-sectional cohorts, ER and GLUT1 (within-sample and between-sample divergence, respectively; see Immunohistochemistry characterization methods section). GLUT1 intensity was different between longitudinal cohorts ( Fig. 5A , p = 0.04, Kruskal-Wallis Rank Sum), like in the cross-sectional study ( Fig. 3A ), with progressors having a generally higher intensity than nonprogressor cohorts, but the pairwise differences were not statistically significant (vs. nonrecurrents p = 0.06, vs. recurrents p = 0.07). ER intensity ( Fig. 5B ) was higher in ER+ progressors ( p = 0.02) and recurrents ( p = 0.03) than in nonrecurrents (Dunn’s test corrected for multiple tests with the Holm-Šidák adjustment). This new pattern was not found in the cross-sectional study, and the difference between progressors and nonrecurrents is robust to ER status stratification ( Supplementary Fig. S5 ).

Distribution of mean normalized intensities (MIS) per patient (see Methods ) in the three longitudinal cohorts (Nonrec: nonrecurrents , Rec: recurrents , Prog: progressors ). A : GLUT1 marker, B : ER marker in ER+ patients only. Omnibus test: Kruskal-Wallis Rank Sum, Post-hoc test: Dunn’s test with control for multiple tests using the Holm-Šidák adjustment. P-values shown if adjusted p ≤ 0.1. Interquartile range (vertical line) and median (point) in burgundy. N: number of patients.
We assessed the phenotypic divergence for these two markers using the same methodology as in the cross-sectional study, evaluating ER within-sample divergence and GLUT1 between-sample divergence, but neither showed a statistically significant difference between longitudinal cohorts ( Supplementary Fig. S6 ).
We tested if our genetic and phenotypic markers were independently associated with the time to non-invasive recurrence or progression using Cox regression analyses. Additionally, alternative clinical outcomes (recurrence [including progression] and progression with non-invasive recurrents right-censored) can be found in the supplementary materials ( Supplementary Figs. S7 - S8 , S10 , Supplementary Tables S7 - S8 ).
Time to non-invasive recurrence was associated with divergences: SNV ( p = 0.024), within-sample ER ( p = 0.026), and CNA ( p = 0.038), while time to progression was primarily associated with totals: SNV burden ( p < 0.0001), ER intensity ( p = 0.025), GLUT1 intensity ( p = 0.027), and CNA burden ( p = 0.045), but also CNA divergence ( p = 0.025) ( Supplementary Tables S9 - S10 , Wald test). The association between SNV burden and progression was the only one that survived multiple-test correction ( Supplementary Tables S7 - S8 , progression adjusted p < 0.0001, Holm correction). Accordingly, we show the capability of this genetic measurement to stratify patients’ non-invasive-recurrence-free ( Fig. 6A ) and progression-free ( Fig. 6B ) survival by splitting patients into low and high SNV burden categories and comparing their event-free survival curves. The Kaplan-Meier plots show differences in the event-free survival curves, with median times to event that differ between groups 100 months for non-invasive recurrence ( Fig. 6A , p = 0.026) and 57 months for progression ( Fig 6B , p < 0.0001, Log-rank test).

Kaplan-Meier plots of stratified patients. A : Non-invasive-recurrence-free survival. B : Progression-free survival. SNV burden thresholds maximize Youden’s J statistic of the outcomes (17 SNVs for non-invasive recurrence and 21 for progression). Log-rank test. The table below the Kaplan-Meier plot shows the number of samples at risk at different times.
Finally, we integrated 18 clinical covariates ( Supplementary Table S4 ) with our genetic and phenotypic measurements to develop comprehensive models of DCIS non-invasive recurrence and progression. Proportional hazard regressions built with variables selected using LASSO contained three significant variables for non-invasive recurrence ( Fig. 7A , treatment option p < 0.001, ER status p = 0.003, and SNV divergence p = 0.018, Wald test) and two for progression ( Fig. 7C , surgical margin p = 0.017, and SNV burden p = 0.004, Wald test), and event-free survival curves of patients stratified using their relative risk were highly significant, with median time to events that differ between groups in 123 months for non-invasive recurrence ( Fig. 7B ) and > 69 months for progression ( Fig. 7D ). An alternative parameterization of the surgical margin as a 2mm threshold showed very similar results ( Supplementary Fig. S9 , p = 0.048, Wald test). The associations with the treatment option and ER status were repeatable without using covariate imputation ( Supplementary Fig. S10 ), while the surgical margin association was only robust when not excluding recurrent patients ( Supplementary Figs. S11 - S12 ). No other significant variables in these models were imputed.
Evolutionary measurements summarize the results of complex evolutionary dynamics, and equivalent observations may result from very different evolutionary scenarios. For example, both a low mutation rate under neutral evolution and a hard selective sweep can generate low divergence of high allele-frequency mutations. Divergence also has multiple scales, and multiple evolutionary processes may affect scales differently or even in opposite directions. Clonal expansion may reduce within-sample divergence but increase between-sample divergence. Intra-tumor heterogeneity provides the fuel for natural selection, but it is not clear what form of intra-tumor heterogeneity (genetic, epigenetic, or phenotypic) is most relevant to the clinical outcomes of a particular tumor, and it is not clear how best to measure it ( 18 ).
The improved efficacy of preventive screenings provided the ability to identify many tumors in the earliest phases of their evolution, demanding the development of new approaches to stratify the risk to these patients to avoid over- and undertreatment. However, every neoplasm develops a unique set of alterations through somatic evolution ( 18 ), making it unlikely that any given set of molecular markers will be universally applicable, even within a given cancer type. In contrast, measures of the evolvability of a neoplasm, such as the number of mutations and measures of intra-tumor heterogeneity, may be universal biomarkers that predict neoplastic progression in many different types of cancers and pre-cancers ( 9 , 10 , 15 ). By taking two spatially distinct samples for each primary pre-cancer, we measured genetic and phenotypic divergence within and between samples, and their relationship with two key clinical processes: 1) recurrence of precancer following treatment and 2) progression of precancer to invasive cancer.
DCIS recurrence and progression are different biological processes
Based on our results, progression from DCIS to invasive breast cancer appears to be a qualitatively and biologically different process from recurrence of DCIS. We had assumed that progression to invasion first requires recurrence of the DCIS and so expected that the factors that predicted recurrence would also predict progression. We were surprised that there was no overlap in their multivariate models ( Fig. 7 ).
Among all genetic and phenotypic variables, SNV burden, as measured with our previously released software ITHE ( 20 ), was the variable that showed the largest differences between the patients that did not recur, the patients that recurred with DCIS, and the patients that progressed to IDC. SNV burden also had the strongest independent association with time to progression and was an essential component of its best multivariate model. The lpWGS CNA burden from the same samples corroborated this finding with higher p-values. Theoretically, this increase in mutation burden may result from an increase in mutation rate, evolutionary time, or self-renewing cell population size. However, due to limitations in detecting variants at low allele frequency, measured mutation burdens are biased towards high allele frequency mutations and are thus most sensitive to early increases in mutation rates or the selective evolutionary forces that drive clonal expansion ( 35 , 36 ). This bias is especially true when using our program ITHE since, by design, it maximizes specificity in exchange for a lower sensitivity for low-frequency mutations in a sample. For these reasons, we do not necessarily expect SNV burden measured differently to show the associations found here.
Progression was also associated with two other magnitude measurements (i.e., totals: ER and GLUT1 intensities) but did not provide enough additional information over the SNV burden to be included as significant variables in the best multivariate model, which also included the size of the surgical margin as a significant predictor.
Previous studies have shown that surgical margins are clinically important in reducing the risk of ipsilateral breast tumor recurrence after breast-conserving surgery ( 37 , 38 ). Positive margins (i.e., DCIS at the edge of the resected tissue) clearly increase recurrence risk, but patients with positive margins were excluded from our study. Instead, we analyzed how the size of the negative margins associate with the clinical outcome. The evidence for this association is mixed in the literature ( 37 , 39 ), but current consensus guidelines consider margins >2mm adequate. Notably, these studies do not typically differentiate recurrence of DCIS from progression to invasive disease in their endpoints, as we did here. We found that the size of the surgical margins was one of the strongest predictors of progression but was not a statistically significant predictor of recurrence with DCIS, neither in the selected multivariate model nor in isolation. This negative result may be due to a type II error, but even if such an association exists, it is likely to be weaker than that observed for progression. We hypothesize that a micro-invasive phenotype could reduce the probability of obtaining large surgical margins, or a phenotype that makes DCIS cells more independent could allow small clusters of cells left over during surgical treatment to survive and further progress to invasive disease more readily. This finding highlights the importance of segregating non-invasive recurrence from progression and how associations with recurrence (of any kind) are primarily a composite of the associations with non-invasive recurrence and progression ( Supplementary Fig. S8A ). We confirmed our results using the consensus guideline >2mm threshold instead of treating surgical margins as a continuous variable, obtaining equivalent though weaker results. This observation shows prognostic information in the size of the surgical margin. The fact that all associations with progression held independently of whether we excluded recurrent patients or right-censored them at the time of DCIS recurrence ( Supplementary Fig. S8C - D ) shows their robustness and adds evidence towards non-invasive recurrence and progression being qualitatively different phenomena.
In contrast, time to non-invasive recurrence was associated with the extent of genetic divergence of SNVs between the two assayed regions of DCIS. We could not corroborate this finding with CNA divergence, which followed the opposite trend but was also correlated to time to recurrence in the univariate models. The true (i.e., known without error) amount of genetic divergence measured using different mutation types should yield equivalent results if large enough mutational burdens of both types are accumulated. A few estimation biases may explain the discordance we observed between SNV and CNA divergences. A low CNA burden may increase the estimated divergence due to a higher false positive rate in the segmentation process without a broad range of true relative intensity values. In fact, CNA burden and CNA divergence were moderately anticorrelated across the study (ρ = −0.36, p < 0.001), and this anticorrelation was driven by the cohort with the lowest CNA burden. High within-sample heterogeneity is also expected to reduce the accuracy of between-sample divergence estimates and lead to the underestimation of the mutation burden. Low SNV burden also leads to missing data in SNV divergence estimates since divergence cannot be calculated accurately with few alterations. ER divergence followed the same direction as CNA divergence, with greater divergence associated with a lower risk of recurrence, but SNV divergence followed the opposite trend. These divergences were the only three measurements associated with time to non-invasive recurrence in the univariate analyses ( Supplementary Table S9 ). Non-invasive recurrence is associated exclusively with divergence statistics, while progression was primarily associated with totals (SNV burden and mean GLUT1 intensity). Intratumor heterogeneity can arise from an increase in the amount of evolution (same mechanisms as mutation burden above) but also with diversifying selection, and we have previously associated it with poor prognosis in other pre-cancers ( 9 ).
Non-invasive recurrence was also associated with the type of DCIS treatment and estrogen-negative status. The fact that patients treated with lumpectomy alone were more likely to recur than those treated with lumpectomy and radiation or mastectomy has been well described. Adjuvant radiation therapy has been previously shown to reduce the risk of recurrence ( 40 ), and after mastectomy, patients are no longer screened using mammograms, making it unlikely that asymptomatic noninvasive recurrences would be detected. The association between recurrence and ER status may be unsurprising since patients with ER+ breast cancers have better prognoses than ER- ones ( 41 , 42 ). However, its association with DCIS recurrence is unclear ( 43 – 45 ), and the balance of evidence points against it ( 46 ). As for surgical margins, most studies are limited by not differentiating between recurrence and progression endpoints. At least one of the studies that made this distinction ( 43 ) found a decrease in non-invasive, but not in invasive recurrences in ER+ patients, which is consistent with our results. Different endpoints may partially explain the mixed evidence on the association between ER and DCIS recurrence and progression.
Functional genetic analysis also showed a difference between the three cohorts, particularly between those DCIS that recurred compared to those that progressed. DCIS that will recur without invasion shows enrichment of mutations in genes involved in the TAS2R signaling network. The activation of these genes determines a pro-apoptotic, anti-proliferative, and anti-migratory response action in highly metastatic breast cancer cell lines ( 47 ). These genes also appear to be involved in the regulation of apoptosis in head and neck squamous cell carcinoma, and their impairment could favor the survival of cancer cells ( 48 ). On the other hand, DCIS that will progress to invasion demonstrates a broader variety of biological processes and pathways involved, such as hypoxia response, insulin/IGF, endothelin, hedgehog, p53, and PI3 kinase signaling pathways. These biological processes are typically altered in various types of cancer and also show an enrichment of mutations in genes involved in the reorganization of the cytoskeleton. The ability to metastasize outside the mammary gland and to relapse observed in these patients is supported by mutations in those pathways.
Synchronous DCIS is not a good model for DCIS progression
Cross-sectional studies are much less resource-intensive, faster, and simpler to conduct than longitudinal cohort studies. If synchronous DCIS (adjacent to IDC) was a good model for primary DCIS that later progressed to IDC, cross-sectional studies could be more readily employed as relevant surrogates for cancer progression. However, our results show this is not possible for our purpose, and in fact, synchronous DCIS shares more similarities with DCIS that will recur as DCIS than with DCIS that will progress.
The pure DCIS samples in our cross-sectional study are equivalent to a mixture of samples from the three cohorts in our longitudinal study since their future outcomes are not considered. Thus, characteristics associated with clinical outcomes are expected to be mixed in the cross-sectional study. We found that DCIS adjacent to IDC showed increased divergence, which may result from divergent evolution facilitated by longer evolutionary times, the interaction with IDC, or an intrinsic characteristic of early-progression DCIS. If we assume that IDC originates from DCIS (stepwise progression model), synchronous DCIS samples are (on average) evolutionarily older than pure DCIS samples, representing a later evolutionary stage than samples from either study. In this case, the cross-sectional study would reveal differences between early and late DCIS. Alternatively, if we assume that an early progression model is also possible (i.e., born to be bad ( 49 )), synchronous DCIS would be enriched with this DCIS sub-type. In this case, the cross-sectional study would show evolutionary characteristics that distinguish those DCIS fated for invasive progression. Additionally, the presence of IDC near synchronous DCIS may also alter its characteristics, modifying its environment systemically (e.g., immune response) and locally (e.g., microenvironment and cell composition through cell migration).
The higher between-sample genetic divergence we found in synchronous DCIS compared to pure DCIS aligns better with stepwise DCIS progression, in which late DCIS would have had more evolutionary time to undergo divergent evolution. Under the early progression model, this may be an intrinsic characteristic of such a DCIS subtype that could facilitate the rapid invasion of nearby tissues. Most (75%) markers with significantly different between-sample divergences showed higher divergence in synchronous DCIS, and all markers with significantly different within-sample divergences showed the opposite trend. These results are concordant with the genetic results and our expectations under a stepwise progression model but did not survive multiple-test correction.
Integrating the results with clonal evolution in neoplastic progression
The two observational studies we conducted here are complementary and together improve our understanding of the evolutionary process leading to DCIS progression and recurrence. We find that primary DCIS that will progress to IDC is more genetically and phenotypically evolved, with higher SNV and CNA burden and more aggressive phenotypes, both metabolically and with respect to its estrogen sensitivity. At least one selective sweep is likely a part of their evolutionary history, which would reduce genetic divergence in the tumor. Higher cell motility could also reduce between-sample heterogeneity. Surgical margins show the strongest association with progression, suggesting that there may be features of the growth pattern of these lesions that make it more difficult to completely excise surgically. In contrast, DCIS recurrence may be primarily enabled by suboptimal clinical management. The few evolutionary features associated with DCIS recurrence suggest an increased accumulation of evolutionary changes in those lesions compared to those that do not recur, which nevertheless do not attain the degree of divergence necessary for invasive progression. In aggregate, the evolutionary history of DCIS recurrences may lack the strong selective sweeps that may be necessary conditions to invade other tissues successfully. DCIS adjacent to IDC shows increased divergence, which may result from divergent evolution facilitated by longer evolutionary times, the interaction with IDC, or an intrinsic characteristic of early-progression DCIS (i.e., born to be bad).
Conclusions
In summary, the evolutionary and clinical measures that predict the recurrence of DCIS differ from those that predict progression to IDC. Furthermore, DCIS adjacent to concurrent invasive cancer appears to be distinct from DCIS that will progress to invasive cancer over time. These findings suggest that the biological dynamics that make DCIS likely to recur differ from those that make it likely to progress, and those dynamics interact differently with our clinical interventions. These insights have the potential to improve both risk stratification and individualized patient management for high-risk DCIS.
Significance
Evolutionary measures of breast pre-cancers associate with local recurrence after surgery, as well as progression to cancer. Recurrence and progression are different biological processes impacted differently by clinical interventions.
Supplementary Material
Supplement 1, acknowledgments.
We thank the Research Computing at Arizona State University for providing HPC ( 50 ) and storage resources that have contributed to the research results reported here. This work is supported in part by NIH grants U54 CA217376, U2C CA233254, R21 CA257980, and R01 CA140657, as well as CDMRP Breast Cancer Research Program Award BC132057 and the Arizona Biomedical Research Commission grant ADHS18-198847. The findings, opinions, and recommendations expressed here are those of the authors and not necessarily those of the universities where the research was performed or the National Institutes of Health.
Conflict of Interest Statement: The authors declare no potential conflicts of interest
- Open access
- Published: 30 August 2024
Hypertension subtypes and adverse maternal and perinatal outcomes - a retrospective population-based cohort study
- Daniel Perejón 1 , 2 , 3 ,
- Anna Bardalet 4 ,
- Iñaki Gascó 5 ,
- Júlia Siscart 1 , 2 , 6 ,
- Maria Catalina Serna 1 , 2 , 7 &
- Míriam Orós 1 , 2 , 8
BMC Pregnancy and Childbirth volume 24 , Article number: 568 ( 2024 ) Cite this article
Metrics details
This study aims to examine risk of adverse pregnancy outcomes and mothers’ characteristics in patients with chronic hypertension, gestational hypertension and preeclampsia.
The study included all births born from women aged 15–45 years, in Lleida, Spain from 2012 to 2018. Pregnancy outcomes were retrieved by regional administrative databases. Logistic regression analysis was used to calculate adjusted odds ratios (OR) (OR 95% CI) for maternal characteristics or neonatal outcomes.
Among 17,177 pregnant women, different types of hypertension present varying risks for both the mother and fetus. There is an increased risk of cesarean section in patients with preeclampsia (OR 2.04, 95% CI: 1.43–2.88). For the newborn, a higher risk of preterm birth is associated with maternal chronic hypertension (OR 3.09, 95% CI: 1.91–4.83) and preeclampsia (OR 5.07, 95% CI: 3.28–7.65). Additionally, there is a higher risk of low birth weight in cases of maternal chronic hypertension (OR 3.2, 95% CI: 2.04–4.88), preeclampsia (OR 5.07, 95% CI: 3.34–7.52), and gestational hypertension (OR 2.72, 95% CI: 1.49–4.68). Furthermore, only newborns of patients with preeclampsia had a higher risk of an Apgar score lower than 7 in the first minute (OR 2.95, 95% CI: 1.45–5.38).
Conclusions
In the study population adjusted for body weight, the different types of hypertension represent different risks in the mother and foetus. These complications were mostly associated with preeclampsia.
Peer Review reports
Introduction
Hypertensive disorders in pregnancy (HDP) are significant contributors to elevated maternal morbidity and mortality rates [ 1 , 2 ], along with neonatal morbidity [ 1 , 2 ], as well as neonatal morbidity. HDP refers to gestational hypertension, preeclampsia and eclampsia, chronic hypertension complicated with preeclampsia, and chronic hypertension [ 3 , 4 ]. According to the International Society for the Study of Hypertension in Pregnancy in 2021, HDP is classified into chronic hypertension, which exists or is diagnosed before 20 weeks’ gestation, and de novo hypertension, which typically occurs from 20 weeks’ gestation onwards. This second one has many manifestations including hypertension alone, known as gestational hypertension; pre-eclampsia (PE), hypertension with proteinuria and maternal organ dysfunction (haematological, liver, renal and neurological) and eclampsia, characterised by seizures [ 5 , 6 ].
Most guidelines around the world agree on the definition of hypertension in pregnancy, consisting in blood pressure (BP) ≥ 140/90 mmHg. At the same time, there is variability in the threshold for initiating antihypertensive treatment attributable to uncertainty about the maternal benefits of lowering BP and the potential foetal risks from reductions in utero-placental circulation and in utero exposure to drugs [ 7 ].
Hypertension in pregnancy is associated with an increased risk of placental abruption, intrauterine growth restriction, preterm birth, renal failure, postpartum haemorrhage, perinatal and maternal death and newborn morbidity [ 8 , 9 , 10 ]. In this sense, it has been estimated that hypertension during pregnancy is one of the main causes of maternal and foetal morbidity and mortality in the world [ 11 ].
Therefore, the aim of this study is to determine the difference in pregnancy outcomes in women with chronic hypertension, gestational hypertension and preeclampsia compared to women with normal pregnancies using populations data.
Materials and methods
Study design and data collection.
A retrospective observational cohort study was conducted among pregnant women in the health region of Lleida from 2012 to 2018.
The data of women who had given birth at the Arnau de Vilanova Hospital between January 1st, 2012 and December 31st, 2018 were obtained through the (“Conjunt Minim de Base de Dades”) CMBD database. Data of all the eligible patients assigned to a primary care unit derived from the computerized clinical history database E-CAP of the Catalan Health Institute; and data from Social Security prescriptions obtained from the database of the ServeiCatalà de Salut.
This article is part of the Iler Pregnancy project, a retrospective cohort study conducted in Lleida with the aim of evaluating the prevalence of chronic pathologies in pregnancy (hypothyroidism, depression, diabetes mellitus and obesity) and therapeutic adherence to prescribed drugs [ 12 , 13 ].
Study population
Women who have had a birth at the Arnau de Vilanova University Hospital in Lleida between January 1st, 2012, and December 31st, 2018, were included in the study. Women who did not belong to Lleida health region were excluded. To evaluate the representativeness of the sample, we calculated the percentage of pregnant women studied compared to the total of pregnant women in the health region of Lleida. Data was obtained from the database of “Instituto Statistics of Catalonia” (Idescat) (Table 1 ).
Variables recorded
The following variables were recorded: region of origin (Sub Saharan Africa, Latin America, Asia and the Middle East, West Europe, Eastern Europe, and Maghreb) [ 12 ]; body mass index (BMI) which is classified according to low weigh (BMI under 18.5 Kg/m2), overweigh (BMI between 25 and 29.9) and obesity (BMI more than 30); number of pregnancy and twin pregnancy; risk during pregnancy; diabetes and mellitus (code O24.9 at CIE-10.); arterial hypertension (code I10-I16 at l’ICD-10); dyslipidemia (code E78 at l’ICD-10); depression (codes F32.0-F32.9, F33.0-F33.3, F33.8, F33.9, F34.1, or F41.2 at l’ICD-10). Other variables taken into account were risk of the pregnancy; duration of the pregnancy (miscarriage, preterm, term, prolonged); caesarean section; birth weight (< 2500 g = underweight, between 2500 g and 3999 g = normal weight, and ≥ 4000 g = macrosomia), 1-minute and 5-minute Apgar score; and preeclampsia.
Data analysis
We performed a descriptive analysis. Based on delivery status, the cohort was divided into four groups: (1) without HDP, (2) chronic hypertension, (3) gestational hypertension, and (4) preeclampsia. Maternal and neonatal characteristics were compared between groups. Continuous variables were expressed as mean and SD and analyzed using ANOVA with post hoc Scheffé test. Ordinal variables were expressed as median and IQR and analyzed using Kruskal–Wallis H test. Categorical variables were expressed as percentages and analyzed using χ² or Fisher’s exact test. Relative risks of HDP phenotypes and outcomes were estimated using multinomial logistic regression. The model-building process was conducted in two blocks: the first included HDP, and the second included covariates (maternal age, BMI, hypothyroidism, maternal diabetes). Adjusted relative risks were expressed as odds ratios (OR) with 95% confidence intervals (95% CI). The “No hypertension” group served as the reference. Superimposed hypertension was excluded from the analysis.
Ethical aspects
This study was approved by the ethics and clinical research committee at the Institut d’Investigació IDIAP Jordi Gol under the code 19/195-P and carried out in accordance with the principles of the Declaration of Helsinki. Information was obtained from electronic medical records stored in the centralized ECAP database and extracted by the Department of Healthcare Evaluation and Research Management. Therefore, it was not necessary to ask participants to sign an informed consent. The variables in the ECAP database were processed anonymously and with full confidentiality guarantees as established by national Spanish law and Regulation 2016/679 of the European Parliament and of the Council on the protection of natural people regarding the processing of personal data, and to the free movement of such data. Ethics committee of (Idiap Jordi Gol i Gurina) waived the need for informed consent due to retrospective observational cohort study.
The study was started with a sample of 21,375 women who had given birth at the Arnau de Vilanova Hospital in Lleida between 2012 and 2018 (both included). From this sample, 1625 patients were excluded because they did not have a personal identification code (CIP), and 2573 because multiple data from the clinical history was missing. The final study sample included 17,177 patients (Fig. 1 ).

Sample of pregnant women studied
Characteristics of the study population
Among the total sample, 533 (3.10%) women had a diagnosis of high blood pressure. 263 (1.53%) pregnant women were diagnosed of chronic hypertension, 111 (0.65%) pregnant women were diagnosed with gestational hypertension and 134 (0.78%) were diagnosed with preeclampsia. Preeclampsia superimposed on chronic hypertension occurred in 25 cases (0.14%).
It was observed that in pregnant women with chronic arterial hypertension (263), the mean age was 33.9 (± 6.00) years, compared to 30.6 (± 5.85) years in the non-hypertensive population. Regarding BMI, 38.4% of patients with chronic hypertension were obese, 44.1% of patients with gestational hypertension, and 26.6% in case of preeclampsia. However, only 14% of non-hypertensive women were obese. Among maternal complications, the percentage of caesarean sections was 28.5% in the case of chronic hypertension, 30.8% in preeclampsia, 23.4% in gestational hypertension compared to 17% in non-hypertensive women. Among the newborn complications, 7.6% in the case of mothers with preeclampsia had an Apgar score lower than 7 in the first minute compared to 2.4% in the case of mothers without hypertension. Respect preterm birth, 18.3% were preterm in the case of chronic hypertension, 24.4% in preeclampsia, 10.7% in gestational hypertension and 5.5% in the case of absence of maternal hypertension. Low birth weight occurred in 17.6% in cases of chronic hypertension, 14.8% in gestational hypertension, 22.9% in preeclampsia and in 5.6% newborns of mothers without hypertension during pregnancy. In the case of chronic hypertension, it was classified as high or very high risk of pregnancy to a greater extent, affecting 31% and 16.3% respectively (Table 2 ).
In the multivariate analysis of the different phenotypes of hypertension during pregnancy adjusted for the covariates (maternal age, BMI, hypothyroidism, maternal diabetes) showed statistically significant associations in the risk of cesarean section in patients with preeclampsia (OR 2.04 95% CI: 1.43–2.88). For the newborn, higher risk of preterm birth was associated with maternal chronic hypertension (OR 3.09, 95% CI: 1.91–4.83) or preeclampsia (OR 5.07, 95% CI: 3.28–7.65) and higher risk of low birth weight in case of maternal chronic hypertension (OR 3.2, 95% CI: 2.04–4.88), preeclampsia (OR 5.07, 95% CI: 3.34–7.52) and in the case of gestational hypertension (OR 2.72, 95% CI: 1.49–4.68). On the other hand, only newborns of patients with preeclampsia had higher risk of having an Apgar score lower than 7 in the first minute (OR 2.95, 95% CI: 1.45–5.38). Patients classified as high or very high risk were primarily those who presented chronic hypertension (OR 5.45, 95% CI: 2.77–10.22) and followed by preeclampsia (OR 1.21, 95% CI: 0.36–3.22) (Fig. 2 ).
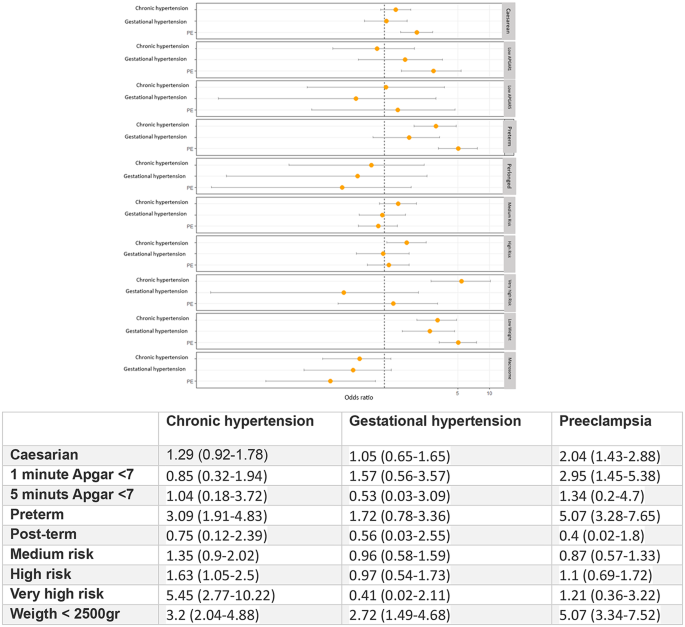
Multivariate analysis of types of hypertension in pregnancy and outcomes in the mother and baby, adjusted for body weight
This study, including 17,177 pregnant women, provides valuable information on the risk factors, prevalence and outcomes of a range of HDP adjusted for body weight, which demonstrates that the different subtypes of hypertension represent different risks to the mother and the foetus. There is an increased risk of caesarean section in patients with preeclampsia (OR 2.04 95% CI: 1.43–2.88). For the newborn, higher risk of preterm birth was associated with maternal chronic hypertension (OR 3.09, 95% CI: 1.91–4.83) or preeclampsia (OR 5.07, 95% CI: 3.28–7.65) and higher risk of low birth weight in case of maternal chronic hypertension (OR 3.2, 95% CI: 2.04–4.88), preeclampsia (OR 5.07, 95% CI: 3.34–7.52) and in the case of gestational hypertension (OR 2.72, 95% CI: 1.49–4.68). On the other hand, only newborns of patients with preeclampsia had higher risk of having an Apgar score lower than 7 in the first minute (OR 2.95, 95% CI: 1.45–5.38). Patients categorized as high or very high risk predominantly include those with chronic hypertension (OR 5.45, 95% CI: 2.77–10.22), followed by those with preeclampsia (OR 1.21, 95% CI: 0.36–3.22).
Analysing risk factors individually, gestational age was significantly higher in patients with chronic hypertension with a median of 33.9 (± 6.19) years of age; being 3 years older in comparison to preeclampsia and non-hypertensive women. BMI average for hypertensive women was 28.8 (± 6.28) and 25.9 (± 5.75) in women with preeclampsia. For the rest of the pregnant women, BMI was 24.8 (± 4.85). In a retrospective cohort study carried out in Southern Spain [ 14 ], it was concluded that overweight and obesity increase the risk of suffering from hypertensive disorders during pregnancy; the risk is significantly higher as BMI increases. In multiple population studies it was identified that obesity increases 2 to 4 times the risk of developing preeclampsia [ 15 , 16 ].
Relationship of chronic hypertension (OR 3.09) and preeclampsia (OR 5.07) with a risk of preterm birth in our study has been observed, as described in other publications. According to Sibai et al., the rates of preterm delivery in a large population of women with chronic hypertension while comparing them with those in a healthy control woman, the overall rates of preterm delivery were significantly higher among women with diabetes mellitus (38%) and hypertension (33.1%) than among control women (13.9%) [ 17 ]. An et al., in a prospective cohort study done in China, after adjusting for potential confounders, observed higher levels of preterm birth in women with gestational hypertension 1.04 (95% CI 0.98 to 1.11) and pre-eclampsia 1.39 (95% CI 1.25 to 1.55), respect control women [ 18 ]. Other medical publications also showed an increased risk of preterm birth in a population with hypertension during pregnancy [ 19 , 20 ].
Delivery methods studies demonstrate higher rate of caesarean section in all women with hypertension: 28.5% in chronic hypertension, 23.4% in gestational hypertension and 30.8% in preeclampsia; compared to 17% in women without hypertension in pregnancy. A systematic review and meta-analysis of hypertension and pregnancy outcomes showed a combined incidence of cesarean section of 41.4% (35.5-47.7%) higher than the rate observed in our study [ 21 ]. Moreover, high incidence of adverse outcomes, were described. Therefore, patient-level analysis should be conducted to assess the reasons for cesarean section to provide and guarantee clear indication in each instance.
Study results are comparable to another study from a maternity hospital in Brazil [ 22 ] that reveals the existence of statistically significant differences between the proportion of c-sections, preterm infants and low birth weight infants for pregnant women with and without hypertensive disorders.
All types of hypertensive disorders were associated with low birth weight. The rate observed for patients with chronic hypertension was 17.6%, 22.9% in patients with preeclampsia, 14.8% in patients with gestational hypertension and 5.6% in women not diagnosed with hypertension.
The study conducted by Fang et al. describes similar results comparing women with and without chronic hypertension; reporting rates of low birth weight among hypertensive mothers for white (16.8%), black (24.4%), and Hispanic (19.5%) populations respectively. Trends were similar for chronic and pregnancy-related hypertension, as well as preeclampsia/eclampsia [ 23 ]. The study completed by Wu et al. evaluates the relationship of stage 1 hypertension detected early in gestation (< 20 weeks) and risks of adverse pregnancy outcomes, stratified by pre-pregnancy BMI. Data indicates that women classified at stage 1a (systolic blood pressure 130–134 mm Hg; diastolic BP, 80–84 mm Hg; or both) and stage 1b hypertension (systolic BP, 135–139 mm Hg; diastolic BP, 85–90 mm Hg; or both) show slightly higher but significant rates and risks of gestational diabetes mellitus, preterm birth, and low birth weight (< 2500 g) in both groups compared with normotensive controls [ 24 ].
Results of this study show that only newborns of patients with preeclampsia had a higher risk of having an Apgar score lower than 7 in the first minute (OR 3.3). However, this was not observed in other hypertensive disorders, where Apgar score was normalizing at 5 min. In a large Chinese population study both maternal hypertension and preeclampsia increased risks for low Apgar score at 1 min (aRR: 1.20, 95%CI: 1.13–1.27; aRR: 1.53, 95%CI: 1.41–1.67, respectively), and for low Apgar score at 5 min (aRR: 1.30, 95%CI: 1.17–1.45; aRR: 1.70, 95%CI: 1.46–1.99, respectively). The risk for neonatal respiratory disorders increased with severity of maternal hypertension [ 25 ]. Moreover, Gu et al. proved that higher diastolic blood pressure was associated with an increased risk of 1-minute Apgar score ≤ 7 when extreme quartiles were compared. However, no significant association was found between systolic blood pressure and 1-minutes or 5-minutes Apgar score ≤ 7, which implies that diastolic blood pressure, has a better prognostic value [ 26 ].
Bronfield et al. [ 27 ]. found in a retrospective study in 14 US states worse outcomes for both mothers and babies in mothers with preeclampsia or superimposed preeclampsia compared to the non-hypertensive population, the population with chronic hypertension also had a higher risk of childbirth premature birth, respiratory distress, low birth weight compared to women without hypertension, but the risk was lower than that of mothers with preeclampsia and, as a last group, women with gestational hypertension had a somewhat higher risk of complications compared to non-hypertensive women but more similar to the healthy population. These data are similar to those reported in our study.
Limitations
The main limitation of this study if the fact of using a retrospective design based on administrative data, thus reducing important information on both maternal and neonatal outcomes. The effect of different antihypertensive treatments on maternal and perinatal outcomes have not been evaluated.
Adequate blood pressure control can modify these adverse outcomes. Minas et cols. [ 28 ] Show that more uncontrollable blood pression patients had superimposed preeclampsia with severe features (54.6% vs. 25.0%; p = 0.01) and preterm delivery (40.9% vs. 10.7%; p = 0.002) than controlled blood pressure patients. The results of CHAP trial [ 29 ] and the meta-analysis carried out by Atta et al. [ 30 ] suggest the beneficence of pharmacologic treatment of mild chronic hypertension during pregnancy to a blood pressure goal below140/90 mm Hg, which is also supported by the Society for Maternal-Fetal Medicine (SMFM) [ 31 ]. Conversely, in our study, we did not analyze the potential complications of eclampsia or HELLP syndrome in a detailed manner, as these conditions are encompassed within the diagnoses of preeclampsia. Furthermore, superimposed preeclampsia was excluded because it involves patients from two distinct groups. Some instances of gestational hypertension may correspond to previously undetected chronic hypertension due to the presence of masked hypertension. This condition has been associated with an increased risk of developing preeclàmpsia [ 32 ].
Finally, another limitation to be considered is the lack of socioeconomic data on the population, which may also influence several factors and health outcomes.
Future research
All types of hypertension have been found to be related to adverse events on pregnancy. This study supports the need to further investigate the pathophysiological knowledge of hypertension in pregnancies to improve the preventive and therapeutic approaches.
Hypertension in pregnancy is associated with higher incidence of adverse pregnancy outcomes. The different types of hypertension represent different risks in the mother and foetus. These complications were mostly associated with preeclampsia. This finding should be interpreted within the limitations of the study.
The use of sensitive diagnostic criteria facilitates solid foundation in epidemiological study, general practise, and clinical research. To address hypertension, Public Health interventions are necessary in addition to clinical management that act at different levels to improve lifestyle habits and early diagnosis before and during pregnancy.
Data availability
The data used in this study are only available for the participating researchers, in accordance with current European and national laws. Thus, the distribution of the data is not allowed. However, researchers from public institutions can request data from SIDIAP.
1, Boulet SL, Platner M, Joseph NT, Campbell A, Williams R, Stanhope KK, Jamieson DJ. Hypertensive Disorders of Pregnancy, Cesarean Delivery, and Severe Maternal Morbidity in an Urban Safety-Net Population. Am J Epidemiol. 2020;189(12):1502–1511. https://doi.org/10.1093/aje/kwaa135 . PMID: 32639535.
Wu P, Chew-Graham CA, Maas AH, Chappell LC, Potts JE, Gulati M, Jordan KP, Mamas MA. Temporal Changes in Hypertensive Disorders of Pregnancy and Impact on Cardiovascular and Obstetric Outcomes. Am J Cardiol. 2020;125(10):1508–1516. doi: 10.1016/j.amjcard.2020.02.029. Epub 2020 Mar 5. PMID: 32273052.
Li F, Qin J, Zhang S, Chen L. Prevalence of hypertensive disorders in pregnancy in China: a systematic review and meta-analysis. Pregnancy Hypertens. 2021;24:13–21. https://doi.org/10.1016/j.preghy.2021.02.001 . Epub 2021 Feb 14. PMID: 33626437.
Article PubMed Google Scholar
ACOG Practice Bulletin No. 202: Gestational Hypertension and Preeclampsia. Obstet Gynecol. 2019;133(1):1. https://doi.org/10.1097/AOG.0000000000003018 . PMID: 30575675.
Magee LA, Brown MA, Hall DR, Gupte S, Hennessy A, Karumanchi SA, Kenny LC, McCarthy F, Myers J, Poon LC, Rana S, Saito S, Staff AC, Tsigas E, von Dadelszen P. The 2021 International Society for the Study of Hypertension in Pregnancy classification, diagnosis & management recommendations for international practice. Pregnancy Hypertens. 2022;27:148–69. Epub 2021 Oct 9. PMID: 35066406.
Corrigan L, O’Farrell A, Moran P, Daly D. Hypertension in pregnancy: prevalence, risk factors and outcomes for women birthing in Ireland. Pregnancy Hypertens. 2021;24:1–6. https://doi.org/10.1016/j.preghy.2021.02.005 . Epub 2021 Feb 14. PMID: 33618054.
American College of Obstetricians and Gynecologists’ Committee on Practice Bulletins—Obstetrics. ACOG Practice Bulletin No. 203: Chronic Hypertension in Pregnancy. Obstet Gynecol. 2019;133(1):e26-e50. https://doi.org/10.1097/AOG.0000000000003020 . PMID: 30575676.
Gestational Hypertension and Preeclampsia: ACOG Practice Bulletin, Number 222. Obstet Gynecol. 2020;135(6):e237-e260. https://doi.org/10.1097/AOG.0000000000003891 . PMID: 32443079.
Steegers EA, von Dadelszen P, Duvekot JJ, Pijnenborg R, Pre-eclampsia. Lancet. 2010;376(9741):631–44. https://doi.org/10.1016/S0140-6736(10)60279-6 . Epub 2010 Jul 2. PMID: 20598363.
Cruz MO, Gao W, Hibbard JU. Obstetrical and perinatal outcomes among women with gestational hypertension, mild preeclampsia, and mild chronic hypertension. Am J Obstet Gynecol. 2011;205(3):260. .e1-260.e9.
Article Google Scholar
Say L, Chou D, Gemmill A, Tunçalp Ö, Moller AB, Daniels J, Gülmezoglu AM, Temmerman M, Alkema L. Global causes of maternal death: a WHO systematic analysis. Lancet Glob Health. 2014;2(6):e323–33. https://doi.org/10.1016/S2214-109X(14)70227-X . Epub 2014 May 5. PMID: 25103301.
12, Orós M, Siscart J, Perejón D, Serna MC, Godoy P, Salinas-Roca B. Ethnic disparities and obesity risk factors in pregnant women: a Retrospective Observational Cohort Study. Nutrients. 2023;15(4):926. https://doi.org/10.3390/nu15040926 . PMID: 36839284; PMCID: PMC9961767.
Siscart J, Orós M, Serna MC, Perejón D, Galván L, Ortega M. Adherence to treatment for hypothyroidism in pregnancy and relationship with thyrotropin control: a retrospective observational cohort study. BMC Pregnancy Childbirth. 2022;22(1):168. https://doi.org/10.1186/s12884-022-04483-8 . PMID: 35232385; PMCID: PMC8886742.
Article CAS PubMed PubMed Central Google Scholar
Fernández Alba JJ, Mesa Páez C, Vilar Sánchez Á, Soto Pazos E, González Macías M, del Serrano Negro C. Overweight and obesity at risk factors for hypertensive states of pregnancy: a retrospective cohort study. Nutr Hosp. 2018;35(4):874–80.
Spradley FT, Palei AC, Granger JP. Increased risk for the development of preeclampsia in obese pregnancies: weighing in on the mechanisms. Am J Physiol Regul Integr Comp Physiol. 2015;309(11):R1326–43. https://doi.org/10.1152/ajpregu.00178.2015 . Epub 2015 Oct 7. PMID: 26447211; PMCID: PMC4698403.
Robillard PY, et al. Increased BMI has a linear association with late-onset preeclampsia: a population-based study. PLoS ONE. 2019;14:e0223888.
Sibai BM, Caritis SN, Hauth JC, MacPherson C, VanDorsten JP, Klebanoff M, Landon M, Paul RH, Meis PJ, Miodovnik M, Dombrowski MP, Thurnau GR, Moawad AH, Roberts J. Preterm delivery in women with pregestational diabetes mellitus or chronic hypertension relative to women with uncomplicated pregnancies. The National institute of Child health and Human Development Maternal- Fetal Medicine Units Network. Am J Obstet Gynecol. 2000;183(6):1520-4. https://doi.org/10.1067/mob.2000.107621 . PMID: 11120521.
An H, Jin M, Li Z, Zhang L, Li H, Zhang Y, Ye R, Li N. Impact of gestational hypertension and pre-eclampsia on preterm birth in China: a large prospective cohort study. BMJ Open. 2022;12(9):e058068. https://doi.org/10.1136/bmjopen-2021-058068 . PMID: 36167382; PMCID: PMC9516080.
Article PubMed PubMed Central Google Scholar
Berhe AK, Ilesanmi AO, Aimakhu CO, et al. Effect of pregnancy induced hypertension on adverse perinatal outcomes in Tigray regional state, Ethiopia: a prospective cohort study. BMC Pregnancy Childbirth. 2019;20:7.
Mulualem G, Wondim A, Woretaw A. The effect of pregnancy induced hypertension and multiple pregnancies on preterm birth in Ethiopia: a systematic review and meta-analysis. BMC Res Notes. 2019;12:91.
Bramham K, Parnell B, Nelson-Piercy C, Seed PT, Poston L, Chappell LC. Chronic hypertension and pregnancy outcomes: systematic review and meta-analysis. BMJ. 2014;348:g2301. https://doi.org/10.1136/bmj.g2301 . PMID: 24735917; PMCID: PMC3988319.
Ramos Filho FL, Antunes CMF. Hypertensive Disorders: Prevalence, Perinatal Outcomes and Cesarean Section Rates in Pregnant Women Hospitalized for Delivery. Rev Bras Ginecol Obstet. 2020;42(11):690–696. doi: 10.1055/s-0040-1714134. Epub 2020 Nov 30. PMID: 33254262; PMCID: PMC10309246.
23, Fang J, Madhavan S, Alderman MH. The influence of maternal hypertension on low birth weight: differences among ethnic populations. Ethn Dis. 1999 Autumn;9(3):369 – 76. PMID: 10600059.
Wu DD, Gao L, Huang O, Ullah K, Guo MX, Liu Y, Zhang J, Chen L, Fan JX, Sheng JZ, Lin XH, Huang HF. Increased Adverse Pregnancy Outcomes Associated With Stage 1 Hypertension in a Low-Risk Cohort: Evidence From 47 874 Cases. Hypertension. 2020;75(3):772–780. doi: 10.1161/HYPERTENSIONAHA.119.14252. Epub 2020 Feb 3. PMID: 32008433.
25, Tian T, Wang L, Ye R, Liu J, Ren A. Maternal hypertension, preeclampsia, and risk of neonatal respiratory disorders in a large-prospective cohort study. Pregnancy Hypertens. 2020;19:131–7. https://doi.org/10.1016/j.preghy.2020.01.006 . Epub 2020 Jan 14. PMID: 31982835.
Gu Y, Shi H, Zeng W, Zheng Y, Yang M, Sun M, Shi H, Gu W. Association between gestational visit-to-visit blood pressure variability and adverse neonatal outcomes. J Clin Hypertens (Greenwich). 2022;24(6):779–88. https://doi.org/10.1111/jch.14500 . Epub 2022 May 14. PMID: 35567772; PMCID: PMC9180330.
Article CAS PubMed Google Scholar
Bromfield SG, Ma Q, DeVries A, Inglis T, Gordon AS. The association between hypertensive disorders during pregnancy and maternal and neonatal outcomes: a retrospective claims analysis. BMC Pregnancy Childbirth. 2023;23(1):514. https://doi.org/10.1186/s12884-023-05818-9 . PMID: 37452285; PMCID: PMC10347833.
Minhas R, Young D, Naseem R, Mueller A, Chinthala S, Perdigao JL, Yeo KJ, Chan SL, Tung A, White JB, Shahul S, Rana S. Association of antepartum blood pressure levels and angiogenic profile among women with chronic hypertension. Pregnancy Hypertens. 2018;14:110–114. doi: 10.1016/j.preghy.2018.09.003. Epub 2018 Sep 5. PMID: 30527096.
Tita AT, Szychowski JM, Boggess K, Dugoff L, Sibai B, Lawrence K, Hughes BL, Bell J, Aagaard K, Edwards RK, Gibson K, Haas DM, Plante L, Metz T, Casey B, Esplin S, Longo S, Hoffman M, Saade GR, Hoppe KK, Foroutan J, Tuuli M, Owens MY, Simhan HN, Frey H, Rosen T, Palatnik A, Baker S, August P, Reddy UM, Kinzler W, Su E, Krishna I, Nguyen N, Norton ME, Skupski D, El-Sayed YY, Ogunyemi D, Galis ZS, Harper L, Ambalavanan N, Geller NL, Oparil S, Cutter GR, Andrews WW. Chronic hypertension and pregnancy (CHAP) Trial Consortium. Treatment for mild chronic hypertension during pregnancy. N Engl J Med. 2022;386(19):1781–92. https://doi.org/10.1056/NEJMoa2201295 . Epub 2022 Apr 2. PMID: 35363951; PMCID: PMC9575330.
Attar A, Hosseinpour A, Moghadami M. The impact of antihypertensive treatment of mild to moderate hypertension during pregnancy on maternal and neonatal outcomes: An updated meta-analysis of randomized controlled trials. Clin Cardiol. 2023;46(5):467–476. doi: 10.1002/clc.24013. Epub 2023 Mar 28. PMID: 369873Society for Maternal-Fetal Medicine. Society for Maternal‐Fetal Medicine statement: antihypertensive therapy for mild chronic hypertension in pregnancy—the chronic hypertension and preg-nancy trial. Am J Obstet Gynecol. 2022;227(2):B24‐B2790; PMCID: PMC10189071.
Society for Maternal-Fetal Medicine. Publications Committee. Electronic address: [email protected]. Society for maternal-fetal Medicine Statement: antihypertensive therapy for mild chronic hypertension in pregnancy-the chronic hypertension and pregnancy trial. Am J Obstet Gynecol. 2022;227(2):B24–7. https://doi.org/10.1016/j.ajog.2022.04.011 . Epub 2022 Apr 19. PMID: 35710594.
Espeche WG, Salazar MR, Minetto J, Leiva Sisnieguez CE, Cerri G, Balbín E, Stavile RN, Carrera Ramos P, Soria A, Santillan C, Grassi F, Torres S, Carbajal HA. Hypertension arising after 20 weeks of gestation: gestational hypertension or masked chronic hypertension? J Hum Hypertens. 2023;37(9):813–7. https://doi.org/10.1038/s41371-022-00767-w . Epub 2022 Oct 12. PMID: 36224324.
Download references
Acknowledgements
The authors would like to acknowledge Dr. Miquel Butí for his valuable contribution and support to design and build the database. Joaquim Sol for his contribution to the statistics analysis, and Gol i Gurina Foundation.
The authors declare no contribution from any organization for the submitted work; no financial relationships with organizations that might have an interest in the submitted work for the previous three years; and no other relationships or activities that could appear to have influenced the submitted work.
Author information
Authors and affiliations.
School of Medicine, Lleida University, Universitat de Lleida, Lleida, Spain
Daniel Perejón, Júlia Siscart, Maria Catalina Serna & Míriam Orós
Institut d’Investigació en Atenció Primària IDIAP Jordi Gol, Catalan Institute of Health, Lleida, Spain
Cervera Health Center, Catalan Institute of Health, Lleida, Spain
Daniel Perejón
Hospital Trueta, Catalan Institute of Health, Anna Bardalet Hospital Trueta, Institut Català de la Salut (ICS), Avda de Francia s/n 17007, Girona, Spain
Anna Bardalet
Hospital Germans Trias, Barcelona, Spain
Iñaki Gascó
Primary Care Centre Seròs, Catalan Institute of Health, Seròs, Spain
Júlia Siscart
Primary Care Centre Eixample, Catalan Institute of Health, Lleida, Spain
Maria Catalina Serna
Cambrils Health Center, Tarragona, Spain
Míriam Orós
You can also search for this author in PubMed Google Scholar
Contributions
AB and DP conceptualized the study, analysed the data, and wrote the first draft of the manuscript; MCS, JS, IG contributed to the design of the study, data management, and manuscript development and review; MO also contributed to the design of the study, and to the creation of data bases and data analysis. All authors read and approved the final manuscript.
Corresponding author
Correspondence to Anna Bardalet .
Ethics declarations
Ethical approval.
This study was approved by the ethics and clinical research committee at the Institut d’Investigació IDIAP Jordi Gol under the code 19/195-P and conducted in accordance with the principles of the Declaration of Helsinki. Information was obtained from electronic medical records stored in the centralized ECAP (computerized clinical history) database and extracted by the Department of Healthcare Evaluation and Research Management. Accordingly, it was not necessary to ask participants for their informed consent. The variables in the ECAP database were processed anonymously and with full confidentiality guarantees as established by Spanish national law and Regulation 2016/679 of the European Parliament and the Council on the protection of natural persons with regard to the processing of personal data, and to the free distribution of such data. The data used in this study are only available for the participating researchers, in accordance with current European and national laws. Thus, the distribution of the data is not allowed. However, researchers from public institutions can request data from SIDIAP. Ethics committee of (Idiap Jordi Gol i Gurina) waived the need for informed consent due to retrospective observational cohort study.
Consent for publication
Not Applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/ .
Reprints and permissions
About this article
Cite this article.
Perejón, D., Bardalet, A., Gascó, I. et al. Hypertension subtypes and adverse maternal and perinatal outcomes - a retrospective population-based cohort study. BMC Pregnancy Childbirth 24 , 568 (2024). https://doi.org/10.1186/s12884-024-06754-y
Download citation
Received : 01 February 2024
Accepted : 12 August 2024
Published : 30 August 2024
DOI : https://doi.org/10.1186/s12884-024-06754-y
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Hypertension
- Pre-eclampsia
- Cesarean section
- Preterm infant
- Low birthweight infant
BMC Pregnancy and Childbirth
ISSN: 1471-2393
- Submission enquiries: [email protected]
- General enquiries: [email protected]
Unignorable influence of chest pain on mood symptoms and prognostic values in coronary artery disease: a cross-sectional study
- Published: 30 August 2024
Cite this article

- Hanxuan Tan 1 na1 ,
- Kun Zeng 1 na1 ,
- Weiya Li 2 na1 ,
- Mingyu Xu 3 ,
- Quanjun Liu 3 &
- Han Yin ORCID: orcid.org/0000-0003-2247-4306 4
Prior researches studying depression and anxiety among individuals with coronary artery disease (CAD) have predominantly concentrated on the connection with clinical and laboratory indicators, disregarding the impact of the cardinal symptom—chest pain. In this cross-sectional study with 561 consecutive CAD inpatients enrolled, the prevalence of mood symptoms/disorder and the influence of chest pain on depression and anxiety symptoms and their prognostic effects in a median follow-up period of 26 months were investigated. The prevalence of having depression and anxiety symptoms reached 37.6% and 27.3%, respectively. Comprehensive analyses revealed that the primary correlated factors for depression were chest pain frequency, age, history of diabetes, and exercise time, and for anxiety were chest pain frequency, chest pain course, and education level. As the common and strongest predictor, chest pain frequency demonstrated a dose-dependent relationship with the risk for mood symptoms. Chest pain frequency and course were not directly associated with prognosis, however impact the prognostic effect of mood symptoms. The association between major adverse cardiovascular events (MACEs) and depression symptoms was primarily observed in patients with a high chest pain frequency, whereas with anxiety was mainly presented in patients with a short chest pain course. For noncardiac rehospitalization, anxiety presented higher predictive value in participants with low chest pain frequencies, while depression was right the opposite. In conclusion, CAD patients with mood symptoms who experience frequent chest pain episodes despite a short course warrant special attention. Enhancing their emotional well-being and addressing chest pain symptoms might potentially yield valuable clinical benefits.
This is a preview of subscription content, log in via an institution to check access.
Access this article
Subscribe and save.
- Get 10 units per month
- Download Article/Chapter or eBook
- 1 Unit = 1 Article or 1 Chapter
- Cancel anytime
Price excludes VAT (USA) Tax calculation will be finalised during checkout.
Instant access to the full article PDF.
Rent this article via DeepDyve
Institutional subscriptions
Data, materials and/or code availability
The data are accessible upon request.
Abbreviations
Coronary angiography
- Coronary artery disease
Generalized Anxiety Disorder Scale
Patient Health Questionnaire
Major adverse cardiovascular event
Allgulander, C. (2016). Anxiety as a risk factor in cardiovascular disease. Current Opinion in Psychiatry, 29 (1), 13–17. https://doi.org/10.1097/yco.0000000000000217
Article PubMed Google Scholar
Bai, B., Yin, H., Guo, L., Ma, H., Wang, H., Liu, F., Liang, Y., Liu, A., & Geng, Q. (2021). Comorbidity of depression and anxiety leads to a poor prognosis following angina pectoris patients: A prospective study. Bmc Psychiatry, 21 (1), 202. https://doi.org/10.1186/s12888-021-03202-5
Article PubMed PubMed Central Google Scholar
Bhatt, K. N., Kalogeropoulos, A. P., Dunbar, S. B., Butler, J., & Georgiopoulou, V. V. (2016). Depression in heart failure: Can PHQ-9 help? International Journal of Cardiology, 221 , 246–250. https://doi.org/10.1016/j.ijcard.2016.07.057
Carney, R. M., & Freedland, K. E. (2017). Depression and coronary heart disease. Nature Reviews. Cardiology, 14 (3), 145–155. https://doi.org/10.1038/nrcardio.2016.181
de Heer, E. W., Palacios, J. E., Adèr, H. J., van Marwijk, H. W. J., Tylee, A., & van der Feltz-Cornelis, C. M. (2020). Chest pain, depression and anxiety in coronary heart disease: Consequence or cause? A prospective clinical study in primary care. Journal of Psychosomatic Research, 129 , 109891. https://doi.org/10.1016/j.jpsychores.2019.109891
Gustad, L. T., Laugsand, L. E., Janszky, I., Dalen, H., & Bjerkeset, O. (2014). Symptoms of anxiety and depression and risk of heart failure: The HUNT study. European Journal of Heart Failure, 16 (8), 861–870. https://doi.org/10.1002/ejhf.133
Harshfield, E. L., Pennells, L., Schwartz, J. E., Willeit, P., Kaptoge, S., Bell, S., Shaffer, J. A., Bolton, T., Spackman, S., Wassertheil-Smoller, S., Kee, F., Amouyel, P., Shea, S. J., Kuller, L. H., Kauhanen, J., van Zutphen, E. M., Blazer, D. G., Krumholz, H., Nietert, P. J., Kromhout, D., Laughlin, G., Berkman, L., Wallace, R. B., Simons, L. A., Dennison, E. M., Barr, E. L. M., Meyer, H. E., Wood, A. M., Danesh, J., Di Angelantonio, E., & Davidson, K. W. (2020). Association between depressive symptoms and Incident Cardiovascular diseases. Jama, 324 (23), 2396–2405. https://doi.org/10.1001/jama.2020.23068
Huysmans, E., Leemans, L., Beckwée, D., Nijs, J., Ickmans, K., Moens, M., Goudman, L., Buyl, R., Putman, K., & Coppieters, I. (2020). The relationship between cognitive and emotional factors and healthcare and medication use in people experiencing pain: A systematic review. J Clin Med, 9 (8). https://doi.org/10.3390/jcm9082486
Jha, M. K., Qamar, A., Vaduganathan, M., Charney, D. S., & Murrough, J. W. (2019). Screening and management of depression in patients with cardiovascular disease: JACC State-of-the-art review. Journal of the American College of Cardiology, 73 (14), 1827–1845. https://doi.org/10.1016/j.jacc.2019.01.041
Johnson, B. D., Shaw, L. J., Pepine, C. J., Reis, S. E., Kelsey, S. F., Sopko, G., Rogers, W. J., Mankad, S., Sharaf, B. L., Bittner, V., & Bairey Merz, C. N. (2006). Persistent chest pain predicts cardiovascular events in women without obstructive coronary artery disease: Results from the NIH-NHLBI-sponsored women’s ischaemia syndrome evaluation (WISE) study. European Heart Journal, 27 (12), 1408–1415. https://doi.org/10.1093/eurheartj/ehl040
Kim, Y., Soffler, M., Paradise, S., Jelani, Q. U., Dziura, J., Sinha, R., & Safdar, B. (2017). Depression is associated with recurrent chest pain with or without coronary artery disease: A prospective cohort study in the emergency department. American Heart Journal, 191 , 47–54. https://doi.org/10.1016/j.ahj.2017.06.003
Kroenke, K., Spitzer, R. L., & Williams, J. B. (2001). The PHQ-9: Validity of a brief depression severity measure. Journal of General Internal Medicine, 16 (9), 606–613. https://doi.org/10.1046/j.1525-1497.2001.016009606.x
Kroenke, K., Spitzer, R. L., & Williams, J. B. (2002). The PHQ-15: Validity of a new measure for evaluating the severity of somatic symptoms. Psychosomatic Medicine, 64 (2), 258–266. https://doi.org/10.1097/00006842-200203000-00008
Kuhlmann, S. L., Arolt, V., Haverkamp, W., Martus, P., Ströhle, A., Waltenberger, J., Rieckmann, N., & Müller-Nordhorn, J. (2019). Prevalence, 12-month prognosis, and clinical management need of depression in coronary heart disease patients: A prospective cohort study. Psychotherapy and Psychosomatics, 88 (5), 300–311. https://doi.org/10.1159/000501502
Li, W., Kai, L., Chang-Ying, W., Li, S., Da-Yi, H., & Rong-Jing, D. (2014). Reliability and validity of GAD-2 and GAD-7 for anxiety screening in cardiovascular disease clinic. Sichuan Mental Health, 3 (27). https://doi.org/10.3969/j.issn.1007-3256.2014.03.002
Lichtlen, P. R., Bargheer, K., & Wenzlaff, P. (1995). Long-term prognosis of patients with anginalike chest pain and normal coronary angiographic findings. Journal of the American College of Cardiology, 25 (5), 1013–1018. https://doi.org/10.1016/0735-1097(94)00519-v
Lichtman, J. H., Bigger, J. T. Jr., Blumenthal, J. A., Frasure-Smith, N., Kaufmann, P. G., Lespérance, F., Mark, D. B., Sheps, D. S., Taylor, C. B., & Froelicher, E. S. (2008). Depression and coronary heart disease: Recommendations for screening, referral, and treatment: A science advisory from the American Heart Association Prevention Committee of the Council on Cardiovascular Nursing, Council on Clinical Cardiology, Council on Epidemiology and Prevention, and Interdisciplinary Council on Quality of Care and Outcomes Research: endorsed by the American Psychiatric Association. Circulation, 118 (17), 1768–1775. https://doi.org/10.1161/circulationaha.108.190769
Löwe, B., Spitzer, R. L., Williams, J. B., Mussell, M., Schellberg, D., & Kroenke, K. (2008). Depression, anxiety and somatization in primary care: Syndrome overlap and functional impairment. General Hospital Psychiatry, 30 (3), 191–199. https://doi.org/10.1016/j.genhosppsych.2008.01.001
McCarron, R. M., Shapiro, B., Rawles, J., & Luo, J. (2021). Depression. Annals of Internal Medicine, 174 (5), Itc65–itc80. https://doi.org/10.7326/aitc202105180
Meyer, T., Hussein, S., Lange, H. W., & Herrmann-Lingen, C. (2015). Anxiety is associated with a reduction in both mortality and major adverse cardiovascular events five years after coronary stenting. Eur J Prev Cardiol, 22 (1), 75–82. https://doi.org/10.1177/2047487313505244
Moazzami, K., Wittbrodt, M. T., Alkhalaf, M., Lima, B. B., Nye, J. A., Mehta, P. K., Quyyumi, A. A., Vaccarino, V., Bremner, J. D., & Shah, A. J. (2020). Association between mental stress-induced inferior frontal cortex activation and angina in coronary artery disease. Circulation: Cardiovascular Imaging, 13 (8), e010710. https://doi.org/10.1161/circimaging.120.010710
Mostofsky, E., Penner, E. A., & Mittleman, M. A. (2014). Outbursts of anger as a trigger of acute cardiovascular events: A systematic review and meta-analysis. European Heart Journal, 35 (21), 1404–1410. https://doi.org/10.1093/eurheartj/ehu033
Najafipour, H., Banivaheb, G., Sabahi, A., Naderi, N., Nasirian, M., & Mirzazadeh, A. (2016). Prevalence of anxiety and depression symptoms and their relationship with other coronary artery disease risk factors: A population-based study on 5900 residents in Southeast Iran. Asian J Psychiatr, 20 , 55–60. https://doi.org/10.1016/j.ajp.2016.01.004
Phan, A., Shufelt, C., & Merz, C. N. (2009). Persistent chest pain and no obstructive coronary artery disease. Jama, 301 (14), 1468–1474. https://doi.org/10.1001/jama.2009.425
Shibeshi, W. A., Young-Xu, Y., & Blatt, C. M. (2007). Anxiety worsens prognosis in patients with coronary artery disease. Journal of the American College of Cardiology, 49 (20), 2021–2027. https://doi.org/10.1016/j.jacc.2007.03.007
Spitzer, R. L., Kroenke, K., Williams, J. B., & Löwe, B. (2006). A brief measure for assessing generalized anxiety disorder: The GAD-7. Archives of Internal Medicine, 166 (10), 1092–1097. https://doi.org/10.1001/archinte.166.10.1092
Thomas, M., Jones, P. G., Arnold, S. V., & Spertus, J. A. (2021). Interpretation of the Seattle Angina Questionnaire as an Outcome measure in clinical trials and clinical care: A review. JAMA Cardiol, 6 (5), 593–599. https://doi.org/10.1001/jamacardio.2020.7478
Valeriani, M., Sestito, A., Le Pera, D., De Armas, L., Infusino, F., Maiese, T., Sgueglia, G. A., Tonali, P. A., Crea, F., Restuccia, D., & Lanza, G. A. (2005). Abnormal cortical pain processing in patients with cardiac syndrome X. European Heart Journal, 26 (10), 975–982. https://doi.org/10.1093/eurheartj/ehi229
van Dijk, M. R., Utens, E. M., Dulfer, K., Al-Qezweny, M. N., van Geuns, R. J., Daemen, J., & van Domburg, R. T. (2016). Depression and anxiety symptoms as predictors of mortality in PCI patients at 10 years of follow-up. Eur J Prev Cardiol, 23 (5), 552–558. https://doi.org/10.1177/2047487315571889
Wu, M., Shen, L., Wang, Q., Liu, L., Lu, S., Jin, J., Dai, Z., & Shu, Z. (2021). Anxiety and depression prevalence and risk factors among patients with cardiovascular diseases in Post-COVID-19 China. Front Public Health, 9 , 758874. https://doi.org/10.3389/fpubh.2021.758874
Yeh, R. W., Tamez, H., Secemsky, E. A., Grantham, J. A., Sapontis, J., Spertus, J. A., Cohen, D. J., Nicholson, W. J., Gosch, K., Jones, P. G., Valsdottir, L. R., Bruckel, J., Lombardi, W. L., & Jaffer, F. A. (2019). Depression and Angina among patients undergoing chronic total occlusion percutaneous coronary intervention: The OPEN-CTO registry. JACC Cardiovasc Interv, 12 (7), 651–658. https://doi.org/10.1016/j.jcin.2018.12.029
Yin, H., Liu, Y., Ma, H., Liu, G., Guo, L., & Geng, Q. (2019). Associations of mood symptoms with NYHA functional classes in angina pectoris patients: A cross-sectional study. Bmc Psychiatry, 19 (1), 85. https://doi.org/10.1186/s12888-019-2061-3
Yin, H., Cheng, X., Liang, Y., Liu, A., Wang, H., Liu, F., Guo, L., Ma, H., & Geng, Q. (2021). High perceived stress may shorten activated partial thromboplastin time and lead to worse clinical outcomes in patients with coronary heart disease. Front Cardiovasc Med, 8 , 769857. https://doi.org/10.3389/fcvm.2021.769857
Zhu, Y., Blumenthal, J. A., Shi, C., Jiang, R., Patel, A., Zhang, A., Yu, X., Gao, R., & Wu, Y. (2018). Sedentary behavior and the risk of depression in patients with acute coronary syndromes. American Journal of Cardiology, 121 (12), 1456–1460. https://doi.org/10.1016/j.amjcard.2018.02.031
Download references
This study was funded by the Natural Science Foundation of Guangdong Province under Grant 2019A1515011224, 2021A1515011118, and 2021A1515011781, and by the High-level Hospital Construction Project of Guangdong Provincial People’s Hospital under Grant DFJH201922, and by Leading Medical Talents in Guangdong Province under Grant KJ012019431.
Author information
Hanxuan Tan, Kun Zeng and Weiya Li contributed equally to this work.
Authors and Affiliations
Wuhan Fourth Hospital, Wuhan, 430032, China
Hanxuan Tan & Kun Zeng
Department of Cardiology, Guangdong Cardiovascular Institute, Guangdong Provincial People’s Hospital, Guangdong Academy of Medical Sciences, Southern Medical University, Guangzhou, 510080, China
School of Medicine, South China University of Technology, Guangzhou, 510006, China
Mingyu Xu & Quanjun Liu
The Second Clinical Medical College, Jinan University (Shenzhen People’s Hospital, The First Affiliated Hospital, Southern University of Science and Technology), Shenzhen, 518020, China
You can also search for this author in PubMed Google Scholar
Contributions
TH and ZK contributed to the conceptualization of the study, literature review, data collection and analysis, and writing the first draft. LW contributed to data analysis, writing and editing. XM and LQ contributed to data collection and analysis, YH contributed to conceptualization of the study, manuscript revision, read and approved the submitted version.
Corresponding author
Correspondence to Han Yin .
Ethics declarations
Ethics approval.
The study was approved by medical ethics committee of GuangdongGeneral Hospital with the following reference number: No.GDREC2017203H. and was performed in line with the principles of the Declaration of Helsinki.
Informed consent was obtained from all individual participants included in the study.
Competing Interests
The authors have no competing interests to declare that are relevant to the content of this article.
Additional information
Publisher’s note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Supplementary Material 1
Rights and permissions.
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Reprints and permissions
About this article
Tan, H., Zeng, K., Li, W. et al. Unignorable influence of chest pain on mood symptoms and prognostic values in coronary artery disease: a cross-sectional study. Curr Psychol (2024). https://doi.org/10.1007/s12144-024-06606-0
Download citation
Accepted : 21 August 2024
Published : 30 August 2024
DOI : https://doi.org/10.1007/s12144-024-06606-0
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Find a journal
- Publish with us
- Track your research

IMAGES
VIDEO
COMMENTS
A case study of multiple-treatments meta-analysis demonstrates that covariates should be considered. A case study of multiple-treatments meta-analysis demonstrates that covariates should be considered. J Clin Epidemiol. 2009 Aug;62 (8):857-64. doi: 10.1016/j.jclinepi.2008.10.001. Epub 2009 Jan 20.
A case study of multiple-treatments meta-analysis demonstrates that covariates should be considered. Georgia Salanti a,b ... Multiple-treatments meta-analysis offers many opportunities, including the abilities to enhance precision, to estimate treatment effects that have not been observed directly, and to rank treatments while fully exploiting ...
Study Design and Setting. We performed multiple-treatments meta-analysis within a Bayesian framework by synthesizing six Cochrane reviews. We explored the compatibility between direct and indirect evidence and adjusted the results using a meta-regression model to take into account differences in the year of randomization across studies.
DOI: 10.1016/j.jclinepi.2008.10.001 Corpus ID: 8052945; A case study of multiple-treatments meta-analysis demonstrates that covariates should be considered. @article{Salanti2009ACS, title={A case study of multiple-treatments meta-analysis demonstrates that covariates should be considered.}, author={Georgia Salanti and Valeria C C Marinho and Julian P. T. Higgins}, journal={Journal of clinical ...
A case study of multiple-treatments meta-analysis demonstrates that covariates should be considered Georgia Salantia,b,*, Valeria Marinhoc, Julian P.T. Higginsa aMRC Biostatistics Unit, Cambridge, UK bClinical and Molecular Epidemiology Unit, Department of Hygiene and Epidemiology, University of Ioannina School of Medicine, Ioannina, Greece cClinical and Diagnostic Oral Sciences, Barts and The ...
A case study of multiple-treatments meta-analysis demonstrates that covariates should be considered. Authors: Salanti G, Marinho V, Higgins JP. ... using as a case study the investigation of the relative effectiveness of four topical fluoride treatments and two control interventions (placebo and no treatment) in preventing dental caries in ...
A case study of multiple-treatments meta-analysis demonstrates that covariates should be considered. Sign in | Create an account. https://orcid.org. Europe PMC. Menu ... API case studies; SOAP web service; Annotations API; OAI service; Bulk downloads; Developers Forum; Help. Help using Europe PMC;
Request PDF | A case study of multiple-treatments meta-analysis demonstrates that covariates should be considered | To illustrate the potential and challenges of the simultaneous analysis of a ...
The multiple-treatments meta-analysis model. Consider a study i that compares toothpaste (T) with rinse (R). The estimated treatment effect in this study is the SMD (toothpaste - rinse), denoted by y TR, i, with estimated variance, s TR, i 2. The estimates are assumed to be normally distributed around the true SMD, δ TR, i: y TR, i ∼ N (δ ...
OBJECTIVE: To illustrate the potential and challenges of the simultaneous analysis of a network of trials, using as a case study the investigation of the relative effectiveness of four topical fluoride treatments and two control interventions (placebo and no treatment) in preventing dental caries in children. STUDY DESIGN AND SETTING: We performed multiple-treatments meta-analysis within a ...
Fig. 5. Assumptions of a meta-regression analysis in which fluoride-control differences change over time (e.g., because of improved oral hygiene; assumed to affect placebo and no-treatment groups). The scenario relates to a specific population under study, and is not assumed to apply across individuals in different studies. - "A case study of multiple-treatments meta-analysis demonstrates that ...
Salanti G, Marinho V, Higgins JP: A case study of multiple-treatments meta-analysis demonstrates that covariates should be considered. J Clin Epidemiol 2009, 62: 857-864.
A case study of multiple-treatments meta-analysis demonstrates that covariates should be considered. G. Salanti, V. Marinho, and J. Higgins. ... using as a case study the investigation of the relative effectiveness of four topical fluoride treatments and two control interventions (placebo and no treatment) in preventing dental caries in ...
We aimed to evaluate current practice when estimating treatment-covariate interactions in IPD meta-analysis, specifically focusing on involvement of additional covariates in the models. We reviewed 100 IPD meta-analyses of randomised trials, published between 2015 and 2020, that assessed at least one treatment-covariate interaction.
1 INTRODUCTION. When many treatments (eg, treatments 1, 2, and 3) exist for the same condition and they form a connected network, network meta-analysis (NMA) can estimate the relative effects of all treatment pairings (eg, 2 vs 1, 3 vs 1, and 3 vs 2) using all direct and indirect evidence. 1-5. The NMA models assume consistency between direct and indirect evidence for the treatment effects. 6 ...
We performed multiple-treatments meta-analysis within a Bayesian framework by synthesizing six Cochrane reviews. We explored the compatibility between direct and indirect evidence and adjusted the results using a meta-regression model to take into account differences in the year of randomization across studies.
A case study of multiple-treatments meta-analysis demonstrates that covariates should be considered. J Clin Epidemiol 2009;62:857-64. ... Autrup H, et al. Cigarette smoking, N-acetyltransferase 2 acetylation status, and bladder cancer risk: a case-series meta-analysis of a gene-environment interaction. Cancer Epidemiol Biomarkers Prev 2000;9: ...
The covariates that were considered for adjustment in both MAIC analyses (i.e. time to CDP-3 and time to CDP-6) included: age, EDSS, duration of MS, duration of SPMS, relapse history, and sex. ... Salanti G, Marinho V, Higgins JP. A case study of multiple-treatments meta-analysis demonstrates that covariates should be considered. J Clin ...
Salanti G, Marinho V, Higgins JP: A case study of multiple-treatments meta-analysis demonstrates that covariates should be considered. J ClinEpidemiol. 2009, 62: 857-864. Google Scholar Jansen JP: Network meta-analysis of individual and aggregate level data. Res Synth Methods. 2012, 3: 177-190. 10.1002/jrsm.1048.
Network meta-analysis, in the context of a systematic review, is a meta-analysis in which multiple treatments (that is, three or more) are being compared using both direct comparisons of interventions within randomized controlled trials and indirect comparisons across trials based on a common comparator. To ensure validity of findings from network meta-analyses, the systematic review must be ...
The ever increasing number of alternative treatment options and the plethora of clinical trials have put systematic reviews and meta-analysis under a new perspective by emphasizing the need to make inferences about competing treatments for the same condition.
Immune checkpoint inhibitors are standard-of-care for the treatment of advanced melanoma, but their use is limited by immune-related adverse events. Proteomic analyses and multiplex cytokine and ...
STUDY DESIGN AND SETTING We performed multiple-treatments meta-analysis within a Bayesian framework by synthesizing six Cochrane reviews. We explored the compatibility between direct and indirect evidence and adjusted the results using a meta-regression model to take into account differences in the year of randomization across studies.
Experimental design. We performed two observational studies (Fig. 1, Table 1) to study DCIS progression.In a cross-sectional study (Fig. 1A), we compared DCIS samples from patients with DCIS only (Pure DCIS, n = 58) versus DCIS samples from patients with synchronous invasive ductal carcinoma (Synchronous DCIS, n = 61).In a longitudinal case-control study (Fig. 1B), we collected samples from ...
Background This study aims to examine risk of adverse pregnancy outcomes and mothers' characteristics in patients with chronic hypertension, gestational hypertension and preeclampsia. Methods The study included all births born from women aged 15-45 years, in Lleida, Spain from 2012 to 2018. Pregnancy outcomes were retrieved by regional administrative databases. Logistic regression analysis ...
Study design and participants. The current study is a post-hoc analysis based on a cross-sectional research conducted at Guangdong Provincial People's Hospital from October 2017 to January 2018, aiming to investigate the prevalence of mood symptoms and their associations with prognosis in patients with CAD (Yin et al., 2019, 2021).The study also included surveys on perceived stress levels ...